4UMJ
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound ibandronic acid molecules. | | 分子名称: | GERANYLTRANSTRANSFERASE, IBANDRONATE, MAGNESIUM ION | | 著者 | Schmidberger, J.W, Schnell, R, Schneider, G. | | 登録日 | 2014-05-18 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2F74
 
 | | Murine MHC class I H-2Db in complex with human b2-microglobulin and LCMV-derived immunodminant peptide gp33 | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | 著者 | Achour, A, Michaelsson, J, Harris, R.A, Ljunggren, H.G, Karre, K, Schneider, G, Sandalova, T. | | 登録日 | 2005-11-30 | | 公開日 | 2006-02-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Basis of the Differential Stability and Receptor Specificity of H-2D(b) in Complex with Murine versus Human beta(2)-Microglobulin.
J.Mol.Biol., 356, 2006
|
|
1RUS
 
 | | CRYSTAL STRUCTURE OF THE BINARY COMPLEX OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE AND ITS PRODUCT, 3-PHOSPHO-D-GLYCERATE | | 分子名称: | 3-PHOSPHOGLYCERIC ACID, RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | 著者 | Lundqvist, T, Schneider, G. | | 登録日 | 1991-10-10 | | 公開日 | 1991-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the binary complex of ribulose-1,5-bisphosphate carboxylase and its product, 3-phospho-D-glycerate.
J.Biol.Chem., 264, 1989
|
|
8R2J
 
 | |
8R2E
 
 | |
8R2B
 
 | |
8R20
 
 | |
2N8D
 
 | |
1ZHB
 
 | | Crystal Structure Of The Murine Class I Major Histocompatibility Complex Of H-2Db, B2-Microglobulin, and a 9-Residue Peptide Derived from rat dopamine beta-monooxigenase | | 分子名称: | 9-mer peptide from Dopamine beta-monooxygenase, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Sandalova, T, Michaelsson, J, Harris, R.A, Odeberg, J, Schneider, G, Karre, K, Achour, A. | | 登録日 | 2005-04-25 | | 公開日 | 2005-06-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A structural basis for CD8+ T cell-dependent recognition of non-homologous peptide ligands: implications for molecular mimicry in autoreactivity
J.Biol.Chem., 280, 2005
|
|
1YBV
 
 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND AN ACTIVE SITE INHIBITOR | | 分子名称: | 5-METHYL-1,2,4-TRIAZOLO[3,4-B]BENZOTHIAZOLE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | 著者 | Andersson, A, Schneider, G, Lindqvist, Y. | | 登録日 | 1996-09-23 | | 公開日 | 1997-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the ternary complex of 1,3,8-trihydroxynaphthalene reductase from Magnaporthe grisea with NADPH and an active-site inhibitor.
Structure, 4, 1996
|
|
3ZQU
 
 | | STRUCTURE OF A PROBABLE AROMATIC ACID DECARBOXYLASE | | 分子名称: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, PROBABLE AROMATIC ACID DECARBOXYLASE, SULFATE ION | | 著者 | Kopec, J, Schnell, R, Schneider, G. | | 登録日 | 2011-06-11 | | 公開日 | 2011-11-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of Pa4019, a Putative Aromatic Acid Decarboxylase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1NGS
 
 | | COMPLEX OF TRANSKETOLASE WITH THIAMIN DIPHOSPHATE, CA2+ AND ACCEPTOR SUBSTRATE ERYTHROSE-4-PHOSPHATE | | 分子名称: | CALCIUM ION, ERYTHOSE-4-PHOSPHATE, THIAMINE DIPHOSPHATE, ... | | 著者 | Nilsson, U, Lindqvist, Y, Schneider, G. | | 登録日 | 1996-09-25 | | 公開日 | 1997-02-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Examination of substrate binding in thiamin diphosphate-dependent transketolase by protein crystallography and site-directed mutagenesis.
J.Biol.Chem., 272, 1997
|
|
5EQU
 
 | | Crystal structure of the epimerase SnoN in complex with Fe3+, alpha ketoglutarate and nogalamycin RO | | 分子名称: | 2-OXOGLUTARIC ACID, FE (III) ION, Nogalamycin RO, ... | | 著者 | Selvaraj, B, Lindqvist, Y, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | 登録日 | 2015-11-13 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EPA
 
 | | Crystal structure of non-heme alpha ketoglutarate dependent carbocyclase SnoK from nogalamycin biosynthesis | | 分子名称: | 2-OXOGLUTARIC ACID, FE (III) ION, MAGNESIUM ION, ... | | 著者 | Selvaraj, B, Lindqvist, Y, Siitonen, V, Niiranen, L, Metsa-Ketela, M, Schneider, G. | | 登録日 | 2015-11-11 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ERL
 
 | | Crystal structure of the epimerase SnoN in complex with Ni2+, succinate and nogalamycin RO | | 分子名称: | NICKEL (II) ION, Nogalamycin RO, SUCCINIC ACID, ... | | 著者 | Selvaraj, B, Lindqvist, Y, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | 登録日 | 2015-11-14 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EP9
 
 | | Crystal structure of the non-heme alpha ketoglutarate dependent epimerase SnoN from nogalamycin biosynthesis | | 分子名称: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | 著者 | Selvaraj, B, Lindqvist, Y, Niiranen, L, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | 登録日 | 2015-11-11 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1TRK
 
 | |
3IHG
 
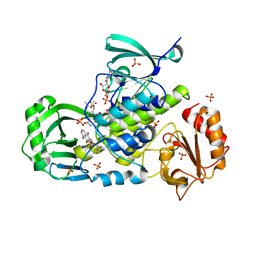 | | Crystal structure of a ternary complex of aklavinone-11 hydroxylase with FAD and aklavinone | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, RdmE, SULFATE ION, ... | | 著者 | Lindqvist, Y, Koskiniemi, H, Jansson, A, Sandalova, T, Schneider, G. | | 登録日 | 2009-07-30 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis.
J.Mol.Biol., 393, 2009
|
|
1ZPD
 
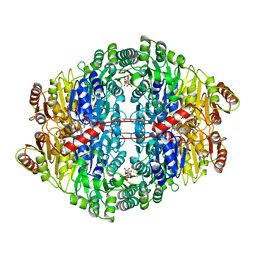 | | PYRUVATE DECARBOXYLASE FROM ZYMOMONAS MOBILIS | | 分子名称: | CITRIC ACID, MAGNESIUM ION, MONO-{4-[(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-AMINO]-2-HYDROXY-3-MERCAPTO-PENT-3-ENYL-PHOSPHONO} ESTER, ... | | 著者 | Lu, G, Dobritzsch, D, Schneider, G. | | 登録日 | 1998-04-17 | | 公開日 | 1999-02-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases.
J.Biol.Chem., 273, 1998
|
|
5EZ7
 
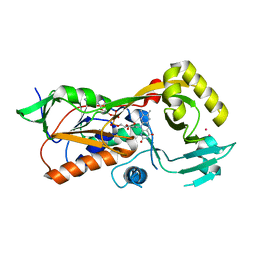 | | Crystal structure of the FAD dependent oxidoreductase PA4991 from Pseudomonas aeruginosa | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, MERCURY (II) ION, flavoenzyme PA4991 | | 著者 | Jacewicz, A, Schnell, R, Lindqvist, Y, Schneider, G. | | 登録日 | 2015-11-26 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the flavoenzyme PA4991 from Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1CNE
 
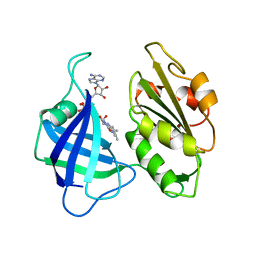 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NITRATE REDUCTASE | | 著者 | Lu, G, Lindqvist, Y, Schneider, G. | | 登録日 | 1995-02-01 | | 公開日 | 1995-04-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
1CNF
 
 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, NITRATE REDUCTASE | | 著者 | Lu, G, Lindqvist, Y, Schneider, G. | | 登録日 | 1995-02-01 | | 公開日 | 1995-04-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
4IWS
 
 | | Putative Aromatic Acid Decarboxylase | | 分子名称: | PA0254, SULFATE ION | | 著者 | Jacewicz, A, Izumi, A, Brunner, K, Schneider, G. | | 登録日 | 2013-01-24 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Insights into the UbiD Protein Family from the Crystal Structure of PA0254 from Pseudomonas aeruginosa.
Plos One, 8, 2013
|
|
1UCW
 
 | |
1XDS
 
 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) and 11-deoxy-beta-rhodomycin (DbrA) | | 分子名称: | 11-DEOXY-BETA-RHODOMYCIN, Protein RdmB, S-ADENOSYLMETHIONINE | | 著者 | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J, Structural Proteomics in Europe (SPINE) | | 登録日 | 2004-09-08 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
