2NA8
 
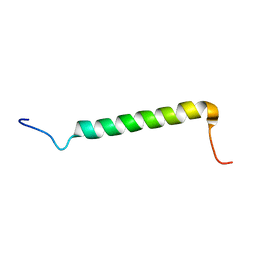 | | Transmembrane Structure of the Cytokine Receptor Common Subunit beta | | Descriptor: | Cytokine receptor common subunit beta | | Authors: | Schmidt, T, Ye, F, Situ, A.J, An, W, Ginsberg, M.H, Ulmer, T.S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Conserved Ectodomain-Transmembrane Domain Linker Motif Tunes the Allosteric Regulation of Cell Surface Receptors.
J.Biol.Chem., 291, 2016
|
|
2N9Y
 
 | |
2NA9
 
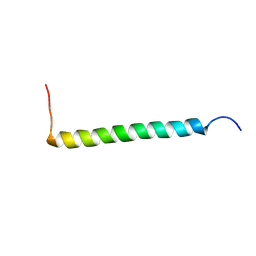 | | Transmembrane Structure of the P441A Mutant of the Cytokine Receptor Common Subunit beta | | Descriptor: | Cytokine receptor common subunit beta | | Authors: | Schmidt, T, Ye, F, Situ, A.J, An, W, Ginsberg, M.H, Ulmer, T.S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Conserved Ectodomain-Transmembrane Domain Linker Motif Tunes the Allosteric Regulation of Cell Surface Receptors.
J.Biol.Chem., 291, 2016
|
|
2L91
 
 | |
1RSU
 
 | |
1RST
 
 | |
6N8C
 
 | | Structure of the Huntingtin tetramer/dimer mixture determined by paramagnetic NMR | | Descriptor: | Huntingtin | | Authors: | Schwieters, C.D, Kotler, S.A, Schmidt, T, Ceccon, A, Ghirlando, R, Libich, D.S, Clore, G.M. | | Deposit date: | 2018-11-29 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Probing initial transient oligomerization events facilitating Huntingtin fibril nucleation at atomic resolution by relaxation-based NMR.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1NY7
 
 | | COWPEA MOSAIC VIRUS (CPMV) | | Descriptor: | COWPEA MOSAIC VIRUS, LARGE (L) SUBUNIT, SMALL (S) SUBUNIT | | Authors: | Lin, T, Chen, Z, Usha, R, Stauffacher, C.V, Dai, J.-B, Schmidt, T, Johnson, J.E. | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Refined Crystal Structure of Cowpea Mosaic Virus at 2.8A Resolution
Virology, 265, 1999
|
|
2UZ1
 
 | | 1.65 Angstrom structure of Benzaldehyde Lyase complexed with 2-methyl- 2,4-pentanediol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZALDEHYDE LYASE, THIAMINE DIPHOSPHATE | | Authors: | Maraite, A, Schmidt, T, Ansorge-Schumacher, M.B, Brzozowski, A.M, Grogan, G. | | Deposit date: | 2007-04-23 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Thdp-Dependent Enzyme Benzaldehyde Lyase Refined to 1.65 A Resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1NQT
 
 | | Crystal structure of bovine Glutamate dehydrogenase-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
4AZ0
 
 | | crystal structure of cathepsin a, complexed with 8a. | | Descriptor: | (S)-3-{[1-(2-Fluoro-phenyl)-5-hydroxy-1H-pyrazole-3-carbonyl]-amino}-3-o-tolyl-propionic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Ruf, S, Buning, C, Schreuder, H, Horstick, G, Linz, W, Olpp, T, Pernerstorfer, J, Hiss, K, Kroll, K, Kannt, A, Kohlmann, M, Linz, D, Huebschle, T, Ruetten, H, Wirth, K, Schmidt, T, Sadowski, T. | | Deposit date: | 2012-06-22 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Beta-Amino Acid Derivatives as Inhibitors of Cathepsin A.
J.Med.Chem., 55, 2012
|
|
4AZ3
 
 | | crystal structure of cathepsin a, complexed with 15a | | Descriptor: | (3S)-3-({[1-(2-fluorophenyl)-5-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-1H-pyrazol-3-yl]carbonyl}amino)-3-(2-methylphenyl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Ruf, S, Buning, C, Schreuder, H, Horstick, G, Linz, W, Olpp, T, Pernerstorfer, J, Hiss, K, Kroll, K, Kannt, A, Kohlmann, M, Linz, D, Huebschle, T, Ruetten, H, Wirth, K, Schmidt, T, Sadowski, T. | | Deposit date: | 2012-06-22 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Novel Beta-Amino Acid Derivatives as Inhibitors of Cathepsin A.
J.Med.Chem., 55, 2012
|
|
1NR1
 
 | | Crystal structure of the R463A mutant of human Glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1NR7
 
 | | Crystal structure of apo bovine glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1L1F
 
 | | Structure of human glutamate dehydrogenase-apo form | | Descriptor: | Glutamate Dehydrogenase 1 | | Authors: | Smith, T.J, Schmidt, T, Fang, J, Wu, J, Siuzdak, G, Stanley, C.A. | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of apo human glutamate dehydrogenase details subunit communication and allostery.
J.Mol.Biol., 318, 2002
|
|
1HWY
 
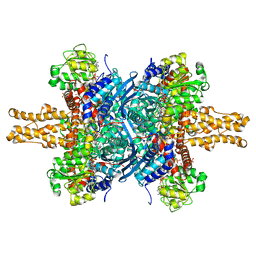 | | BOVINE GLUTAMATE DEHYDROGENASE COMPLEXED WITH NAD AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, GLUTAMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Smith, T.J, Peterson, P.E, Schmidt, T, Fang, J, Stanley, C.A. | | Deposit date: | 2001-01-10 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of bovine glutamate dehydrogenase complexes elucidate the mechanism of purine regulation.
J.Mol.Biol., 307, 2001
|
|
1F15
 
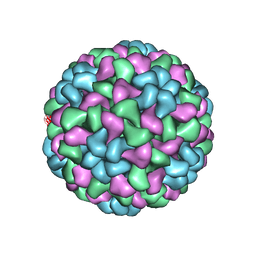 | |
4I4B
 
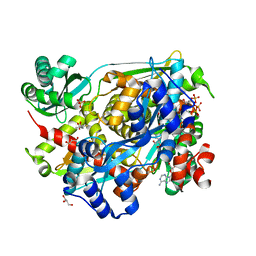 | | HMG-CoA Reductase from Pseudomonas mevalonii complexed with NAD and intermediate hemiacetal form of HMG-CoA | | Descriptor: | (3R,5R,9R,19R,21S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,21-tetrahydroxy-8,8,21-trimethyl-10,14-dioxo-19-sulfanyl-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide, (3S)-3-hydroxy-3-methyl-5-sulfanylpentanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, ... | | Authors: | Stauffacher, C.V, Steussy, C.N, Critchelow, C.J, Schmidt, T, Min, J, Wrensford, L.V, Burgner, J.W, Rodwell, V.W. | | Deposit date: | 2012-11-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
4I64
 
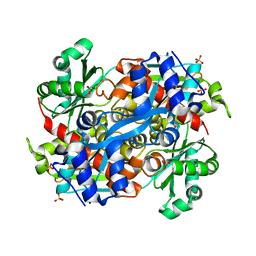 | | 3-hydroxy-3-methylglutaryl Coenzyme A reductase from Pseudomonas mevalonii, a high resolution native structure | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
4I6A
 
 | | 3-hydroxy-3-methylglutaryl (HMG) Coenzyme A reductase from Pseudomonas mevalonii complexed with HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
4I6W
 
 | | 3-hydroxy-3-methylglutaryl (HMG) Coenzyme-A reductase complexed with thiomevalonate | | Descriptor: | (3S)-3-hydroxy-3-methyl-5-sulfanylpentanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, ... | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
4I6Y
 
 | | 3-hydroxy-3-methyl (HMG) Coenzyme A Reductase bound to R-Mevalonate | | Descriptor: | (R)-MEVALONATE, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, ... | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | Deposit date: | 2012-11-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
4I56
 
 | | HMG-CoA reductase from pseudomonas mevalonii complexed with dithio-HMG-coa | | Descriptor: | (3S,5S,9R,21S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,21-tetrahydroxy-8,8,21-trimethyl-10,14-dioxo-19-thioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, ... | | Authors: | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Crichelow, C.J, Rodwell, V.W, Wrensford, L.V, Min, J, Burgner II, J.W. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
1BMV
 
 | | PROTEIN-RNA INTERACTIONS IN AN ICOSAHEDRAL VIRUS AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (ICOSAHEDRAL VIRUS - A DOMAIN), PROTEIN (ICOSAHEDRAL VIRUS - B AND C DOMAIN), RNA (5'-R(*GP*GP*UP*CP*AP*AP*AP*AP*UP*GP*C)-3') | | Authors: | Chen, Z, Stauffacher, C, Li, Y, Schmidt, T, Bomu, W, Kamer, G, Shanks, M, Lomonossoff, G, Johnson, J.E. | | Deposit date: | 1989-10-09 | | Release date: | 1989-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein-RNA interactions in an icosahedral virus at 3.0 A resolution.
Science, 245, 1989
|
|
7BJK
 
 | |
