2MXS
 
 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | Descriptor: | PAROMOMYCIN, RNA (27-MER) | | Authors: | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | Deposit date: | 2015-01-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6VJ7
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with adenosine 5'-(beta,gamma imido)triphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, McGeary, R.P, Mitic, N, Furtado, A, Schulz, B.L, Henry, R.J, Schmidt, S. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops.
Plant Sci., 294, 2020
|
|
2UZX
 
 | | Structure of the human receptor tyrosine kinase Met in complex with the Listeria monocytogenes invasion protein InlB: Crystal form I | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR, INTERNALIN B | | Authors: | Niemann, H.H, Jager, V, Butler, P.J.G, Van Den Heuvel, J, Schmidt, S, Ferraris, D, Gherardi, E, Heinz, D.W. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Human Receptor Tyrosine Kinase met in Complex with the Listeria Invasion Protein Inlb
Cell(Cambridge,Mass.), 130, 2007
|
|
3ZS1
 
 | | Human Myeloperoxidase inactivated by TX5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-METHOXYETHYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
3ZS0
 
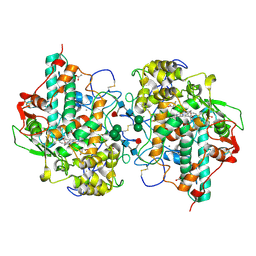 | | Human Myeloperoxidase inactivated by TX2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-FLUOROBENZYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
2UZY
 
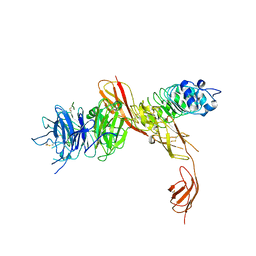 | | Structure of the human receptor tyrosine kinase Met in complex with the Listeria monocytogenes invasion protein inlb: low resolution, Crystal form II | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR, INTERNALIN B | | Authors: | Niemann, H.H, Jager, V, Butler, P.J.G, van den Heuvel, J, Schmidt, S, Ferraris, D, Gherardi, E, Heinz, D.W. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Human Receptor Tyrosine Kinase met in Complex with the Listeria Invasion Protein Inlb
Cell(Cambridge,Mass.), 130, 2007
|
|
6PY9
 
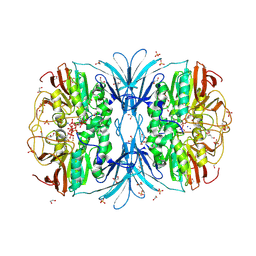 | | Crystal structure of red kidney bean purple acid phosphatase in complex with adenosine diphosphate metavanadate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADP METAVANADATE, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, McGeary, R.P, Mitic, N, Furtado, A, Schulz, B.L, Henry, R.J, Schmidt, S. | | Deposit date: | 2019-07-29 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops.
Plant Sci., 294, 2020
|
|
4RVM
 
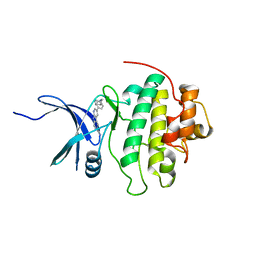 | | CHK1 kinase domain with diazacarbazole compound 19 | | Descriptor: | 3-[4-(piperidin-1-ylmethyl)phenyl]-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Gazzard, L, Blackwood, E, Burton, B, Chapman, K, Chen, H, Crackett, P, Drobnick, J, Ellwood, C, Epler, J, Flagella, M, Goodacre, S, Halladay, J, Hunt, H, Kintz, S, Lyssikatos, J, MacLeod, C, Ramiscal, S, Schmidt, S, Seward, E, Wiesmann, C, Williams, K, Wu, P, Yee, S, Yen, I, Malek, S. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
7BL6
 
 | | 50S-ObgE-GMPPNP particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL2
 
 | | pre-50S-ObgE particle state 1 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL5
 
 | | pre-50S-ObgE particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T, Schmidt, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
3RJQ
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 in complex with C186 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C186 gp120, Llama VHH A12 | | Authors: | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of anti-HIV A12 VHH of llama antibody in complex with C1086 gp120
To be Published
|
|
3R0M
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 | | Descriptor: | Llama VHH A12, SULFATE ION | | Authors: | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | Deposit date: | 2011-03-08 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Single-Headed Immunoglobulins Efficiently Penetrate CD4-Binding Site and Effectively Neutralize HIV-1
To be Published
|
|
8QM0
 
 | | Crystal structure of the pneumococcal Substrate-binding protein AmiA in complex with Peptide 5 | | Descriptor: | ALA-LYS-THR-ILE-LYS-ILE-THR-GLN-THR-ARG, Oligopeptide-binding protein AmiA | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8A42
 
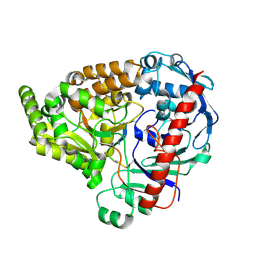 | | Crystal structure of the pneumococcal Substrate-binding protein AmiA in complex with an unknown peptide | | Descriptor: | Oligopeptide-binding protein AmiA, Unknown peptide | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2022-06-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLG
 
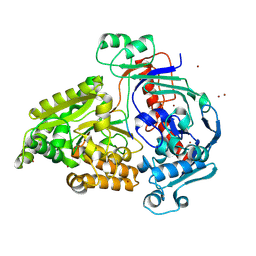 | | Crystal structure of the pneumococcal Substrate-binding protein AliD in closed conformation in complex with Peptide 1 | | Descriptor: | AliD, Peptide 1, ZINC ION | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLH
 
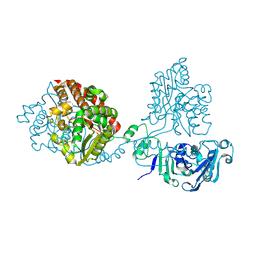 | |
8QLJ
 
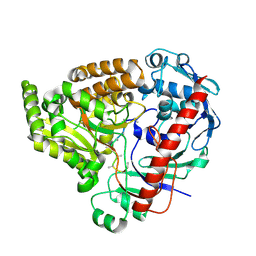 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with an unknown peptide | | Descriptor: | ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, Oligopeptide-binding protein AliB | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLC
 
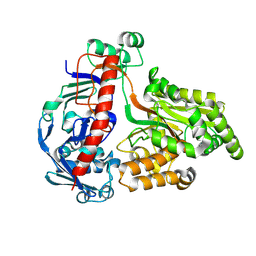 | | Crystal structure of the pneumococcal Substrate-binding protein AliD in open conformation | | Descriptor: | AliD, MAGNESIUM ION | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLK
 
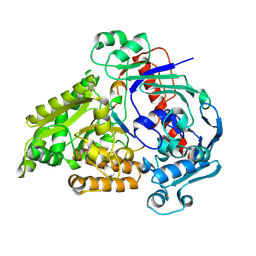 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 2 | | Descriptor: | ALA-ILE-GLN-SER-GLU-LYS-ALA-ARG-LYS-HIS-ASN, Oligopeptide-binding protein AliB | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLM
 
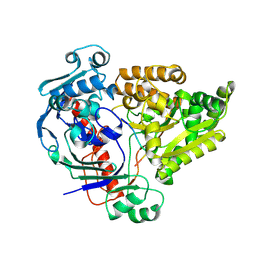 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 3 | | Descriptor: | Oligopeptide-binding protein AliB, PRO-ILE-VAL-GLY-GLY-HIS-GLU-GLY-ALA-GLY-VAL | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8QLV
 
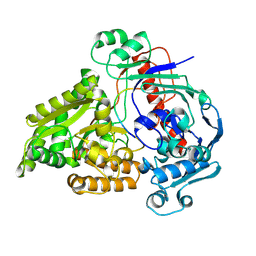 | | Crystal structure of the pneumococcal Substrate-binding protein AliB in complex with Peptide 4 | | Descriptor: | Oligopeptide-binding protein AliB, VAL-MET-VAL-LYS-GLY-PRO-GLY-PRO-GLY-ARG | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
1W6T
 
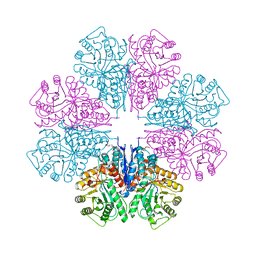 | | Crystal Structure Of Octameric Enolase From Streptococcus pneumoniae | | Descriptor: | ENOLASE, MAGNESIUM ION, NONAETHYLENE GLYCOL | | Authors: | Ehinger, S, Schubert, W.-D, Bergmann, S, Hammerschmidt, S, Heinz, D.W. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasmin(Ogen)-Binding Alpha-Enolase from Streptococcus Pneumoniae: Crystal Structure and Evaluation of Plasmin(Ogen)-Binding Sites
J.Mol.Biol., 343, 2004
|
|
5CYB
 
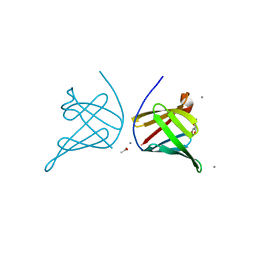 | | Structure of a lipocalin lipoprotein affecting virulence in Streptococcus pneumoniae | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Carrasco-Lopez, C, Abdullah, M.R, Hammerschmidt, S, Hermoso, J.A. | | Deposit date: | 2015-07-30 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the function and structure of a lipocalin lipoprotein affecting virulence in Streptococcus pneumoniae
To Be Published
|
|
2YP6
 
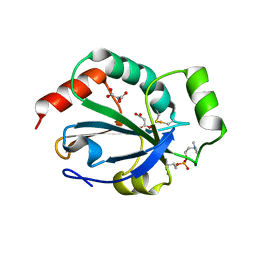 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_1000 (Etrx2) from Streptococcus pneumoniae strain TIGR4 in complex with Cyclofos 3 TM | | Descriptor: | 3-Cyclohexyl-1-Propylphosphocholine, MALONATE ION, THIOREDOXIN FAMILY PROTEIN | | Authors: | Bartual, S.G, Shalek, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Molecular Architecture of Streptococcus Pneumoniae Surface Thioredoxin-Fold Lipoproteins Crucial for Extracellular Oxidative Stress Resistance and Maintenance of Virulence.
Embo Mol.Med., 5, 2013
|
|
