3UU3
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-20' oxidized mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UU4
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in the crystal in a locally-closed conformation (LC1 subtype) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UU5
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-20' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UU8
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TLS
 
 | | The GLIC pentameric Ligand-Gated Ion Channel E19'P mutant in a locally-closed conformation (LC2 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TLV
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-22' oxidized mutant in a locally-closed conformation (LC3 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UUB
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-21' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
9H78
 
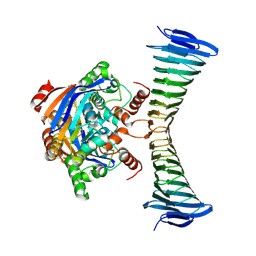 | | [FeS] cluster-loaded SMS complex from M. jannaschii | | Descriptor: | IRON/SULFUR CLUSTER, Iron-sulfur cluster assembly SufBD family protein MJ0034, Uncharacterized ABC transporter ATP-binding protein MJ0035 | | Authors: | Martinez-Carranza, M, Sauguet, L. | | Deposit date: | 2024-10-27 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The ancestral iron-sulfur biogenesis SMS system of Methanocaldococcus jannaschii: a link between geochemistry and microbiology
To Be Published
|
|
8C5Y
 
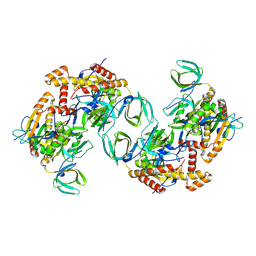 | | RPA tetrameric supercomplex from Pyrococcus abyssi | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Legrand, P, Sauguet, L. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8C5Z
 
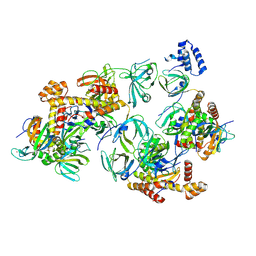 | | RPA tetrameric supercomplex with AROD-OB-1 | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Legrand, P, Sauguet, L. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
4QH5
 
 | | The GLIC pentameric Ligand-Gated Ion Channel (wild-type) crystallized in phosphate buffer | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Delarue, M, Sauguet, L. | | Deposit date: | 2014-05-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of potential modulation sites in the extracellular domain of the prokaryotic pentameric proton-gated ion channel GLIC
Acta Crystallogr.,Sect.D, 2015
|
|
8AAJ
 
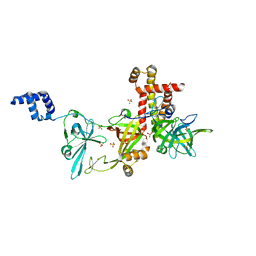 | | Crystal structure of the Pyrococcus abyssi RPA (apo form) | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Legrand, P, Madru, C, Sauguet, L. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8AAS
 
 | | Crystal structure of the Pyrococcus abyssi RPA trimerization core bound to poly-dT20 ssDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, ... | | Authors: | Madru, C, Legrand, P, Sauguet, L. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8AA9
 
 | |
8PPV
 
 | | Intermediate conformer of Pyrococcus abyssi DNA polymerase D (PolD) bound to a primer/template substrate containing three consecutive mismatches | | Descriptor: | DNA (5'-D(*P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*(GS)P*(G7P)P*(PST)P*(PST)P*(PST))-3'), DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA polymerase II small subunit, ... | | Authors: | Betancurt-Anzola, L, Martinez-Carranza, M, Zatopek, K.M, Gardner, A.F, Sauguet, L. | | Deposit date: | 2023-07-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Nat Commun, 14, 2023
|
|
8PPT
 
 | | Pyrococcus abyssi DNA polymerase D (PolD) in its editing mode bound to a primer/template substrate containing a mismatch | | Descriptor: | DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*GP*CP*TP*TP*T)-3'), DNA polymerase II small subunit, ... | | Authors: | Betancurt-Anzola, L, Martinez-Carranza, M, Zatopek, K.M, Gardner, A.F, Sauguet, L. | | Deposit date: | 2023-07-10 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Nat Commun, 14, 2023
|
|
8PPU
 
 | | Pyrococcus abyssi DNA polymerase D (PolD) in its editing mode bound to a primer/template substrate containing three consecutive mismatches | | Descriptor: | DNA (5'-D(P*AP*GP*CP*AP*CP*GP*GP*CP*TP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*GP*CP*CP*GP*AP*GP*CP*CP*GP*TP*(GS)P*(C7R)P*(PST)P*(PST)P*(PST))-3'), DNA polymerase II small subunit, ... | | Authors: | Betancurt-Anzola, L, Martinez-Carranza, M, Zatopek, K.M, Gardner, A.F, Sauguet, L. | | Deposit date: | 2023-07-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Nat Commun, 14, 2023
|
|
8OEJ
 
 | | Extended RPA-DNA nucleoprotein filament | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Sauguet, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (7.96 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8OEL
 
 | | Condensed RPA-DNA nucleoprotein filament | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Sauguet, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (8.24 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
9F28
 
 | |
9F2A
 
 | |
9F26
 
 | | Crystal structure of the PriS_PriL-Rpa2WH ternary complex from P. abyssi | | Descriptor: | DNA primase large subunit PriL, DNA primase small subunit PriS, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, ... | | Authors: | Madru, C, Legrand, P, Haouz, A, Sauguet, L. | | Deposit date: | 2024-04-22 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Communication between DNA polymerases and Replication Protein A within the archaeal replisome.
Nat Commun, 15, 2024
|
|
9F29
 
 | |
6T8H
 
 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T7Y
 
 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
