3B33
 
 | |
2R48
 
 | | Crystal structure of the fructose specific IIB subunit of PTS system from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Phosphotransferase system (PTS) mannose-specific enzyme IIBCA component | | Authors: | Nocek, B, Cuff, M, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-30 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the fructose specific IIB subunit of PTS system from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
2QMA
 
 | | Crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase | | Authors: | Osipiuk, J, Sather, A, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystal structure of glutamate decarboxylase domain of diaminobutyrate-pyruvate transaminase and L-2,4-diaminobutyrate decarboxylase from Vibrio parahaemolyticus.
To be Published
|
|
2QWV
 
 | | Crystal structure of unknown function protein VCA1059 | | Descriptor: | ACETIC ACID, UPF0217 protein VC_A1059 | | Authors: | Chang, C, Sather, A, Moy, S, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of unknown function protein VCA1059.
To be Published
|
|
2R4Q
 
 | | The structure of a domain of fruA from Bacillus subtilis | | Descriptor: | Phosphotransferase system (PTS) fructose-specific enzyme IIABC component | | Authors: | Cuff, M.E, Sather, A, Nocek, B, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of a domain of fruA from Bacillus subtilis.
TO BE PUBLISHED
|
|
3LSO
 
 | |
2R78
 
 | |
3FEU
 
 | |
3C7J
 
 | | Crystal structure of transcriptional regulator (GntR family member) from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | NICKEL (II) ION, Transcriptional regulator, GntR family | | Authors: | Nocek, B, Sather, A, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-07 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of transcriptional regulator (GntR family member) from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
3LK7
 
 | | The Crystal Structure of UDP-N-acetylmuramoylalanine-D-glutamate (MurD) ligase from Streptococcus agalactiae to 1.5A | | Descriptor: | CHLORIDE ION, SULFATE ION, UDP-N-acetylmuramoylalanine--D-glutamate ligase | | Authors: | Stein, A.J, Sather, A, Shakelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of UDP-N-acetylmuramoylalanine-D-glutamate (MurD) ligase from Streptococcus agalactiae to 1.5A
To be Published
|
|
3LW7
 
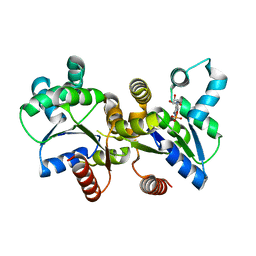 | | The Crystal Structure of an Adenylate kinase-related protein bound to AMP from sulfolobus solfataricus to 2.3A | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylate kinase related protein (AdkA-like) | | Authors: | Stein, A.J, Sather, A, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of an Adenylate kinase-related protein bound to AMP from sulfolobus solfataricus to 2.3A
To be Published
|
|
3FK8
 
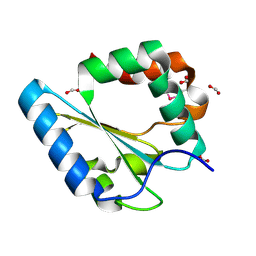 | |
3E0X
 
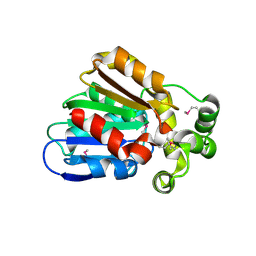 | |
3CK6
 
 | | Crystal structure of ZntB cytoplasmic domain from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | CHLORIDE ION, Putative membrane transport protein | | Authors: | Tan, K, Sather, A, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and electrostatic property of cytoplasmic domain of ZntB transporter.
Protein Sci., 18, 2009
|
|
3G74
 
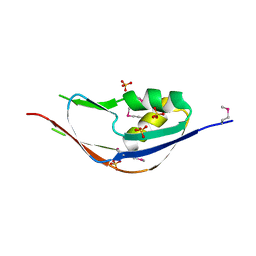 | | Crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560 | | Descriptor: | Protein of unknown function, SULFATE ION | | Authors: | Tan, K, Sather, A, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560
To be Published
|
|
3H04
 
 | | The crystal structure of the protein with unknown function from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | uncharacterized protein | | Authors: | Zhang, R, Tesar, C, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-08 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the protein with unknown function from Staphylococcus aureus subsp. aureus Mu50
To be Published
|
|
3GDW
 
 | | Crystal structure of sigma-54 interaction domain protein from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, Sigma-54 interaction domain protein | | Authors: | Chang, C, Sather, A, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of sigma-54 interaction domain protein from Enterococcus faecalis
To be Published
|
|
3GLV
 
 | | Crystal structure of the lipopolysaccharide core biosynthesis protein from Thermoplasma volcanium GSS1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Lipopolysaccharide core biosynthesis protein, SULFATE ION | | Authors: | Zhang, R, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the lipopolysaccharide core biosynthesis protein from Thermoplasma volcanium GSS1
To be Published
|
|
3IC7
 
 | | Crystal Structure of Putative Transcriptional Regulator of GntR Family from Bacteroides thetaiotaomicron | | Descriptor: | Putative transcriptional regulator | | Authors: | Kim, Y, Sather, A, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.819 Å) | | Cite: | Crystal Structure of Putative Transcriptional Regulator of GntR Family from Bacteroides thetaiotaomicron
To be Published
|
|
3GBY
 
 | | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Uncharacterized protein CT1051 | | Authors: | Fan, Y, Chang, C, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of a protein with unknown function CT1051 from Chlorobium tepidum
To be Published
|
|
3H0K
 
 | |
3IUO
 
 | | The Crystal Structure of the C-terminal domain of the ATP-dependent DNA helicase RecQ from Porphyromonas gingivalis to 1.6A | | Descriptor: | ATP-dependent DNA helicase RecQ, CHLORIDE ION, SODIUM ION | | Authors: | Stein, A.J, Sather, A, Duggan, E, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the C-terminal domain of the ATP-dependent DNA helicase RecQ from Porphyromonas gingivalis to 1.6A
To be Published
|
|
3IH5
 
 | | Crystal Structure of Electron Transfer Flavoprotein alpha-subunit from Bacteroides thetaiotaomicron | | Descriptor: | Electron transfer flavoprotein alpha-subunit, FORMIC ACID, MALONIC ACID, ... | | Authors: | Kim, Y, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Electron Transfer Flavoprotein alpha-subunit from Bacteroides thetaiotaomicron
To be Published
|
|
3H05
 
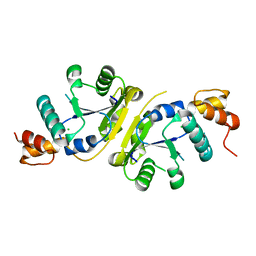 | | The Crystal Structure of a Putative Nicotinate-nucleotide Adenylyltransferase from Vibrio parahaemolyticus | | Descriptor: | CHLORIDE ION, uncharacterized protein VPA0413 | | Authors: | Stein, A.J, Cuff, M.E, Sather, A, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of a Putative Nicotinate-nucleotide Adenylyltransferase from Vibrio parahaemolyticus
To be Published
|
|
3HH1
 
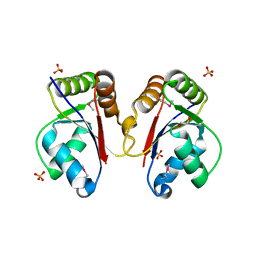 | | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Tetrapyrrole methylase family protein | | Authors: | Cuff, M.E, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Tetrapyrrole methylase family protein domain from Chlorobium tepidum TLS.
TO BE PUBLISHED
|
|
