6LU2
 
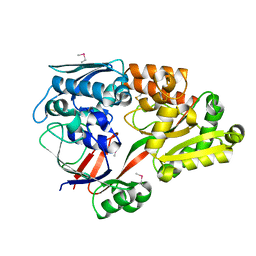 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans | | Descriptor: | Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2020-04-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6LU3
 
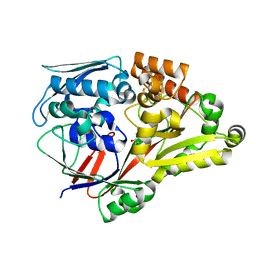 | | Crystal structure of a substrate binding protein from Microbacterium hydrocarbonoxydans complexed with 4-hydroxybenzoate hydrazide | | Descriptor: | 4-oxidanylbenzohydrazide, Substrate binding protein | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
6LU4
 
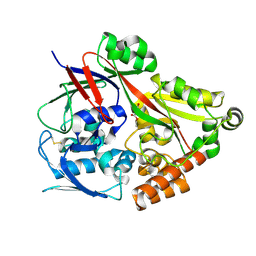 | | Crystal structure of the substrate binding protein from Microbacterium hydrocarbonoxydans complexed with propylparaben | | Descriptor: | Substrate binding protein, propyl 4-hydroxybenzoate | | Authors: | Shimamura, K, Akiyama, T, Yokoyama, K, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-01-25 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of substrate recognition by the substrate binding protein (SBP) of a hydrazide transporter, obtained from Microbacterium hydrocarbonoxydans.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
5H6T
 
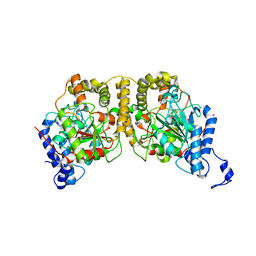 | | Crystal structure of Hydrazidase from Microbacterium sp. strain HM58-2 | | Descriptor: | Amidase | | Authors: | Akiyama, T, Ishii, M, Takuwa, A, Oinuma, K, Sasaki, Y, Takaya, N, Yajima, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5H6S
 
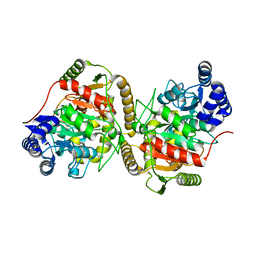 | | Crystal structure of Hydrazidase S179A mutant complexed with a substrate | | Descriptor: | 4-oxidanylbenzohydrazide, Amidase | | Authors: | Akiyama, T, Ishii, M, Takuwa, A, Oinuma, K, Sasaki, Y, Takaya, N, Yajima, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5H0Q
 
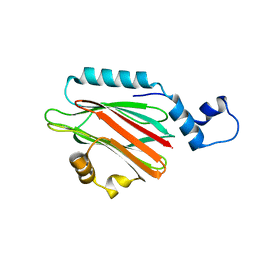 | | Crystal structure of lipid binding protein Nakanori at 1.5A | | Descriptor: | Lipid binding protein | | Authors: | Makino, A, Abe, M, Ishitsuka, R, Murate, M, Kishimoto, T, Sakai, S, Hullin-Matsuda, F, Shimada, Y, Inaba, T, Miyatake, H, Tanaka, H, Kurahashi, A, Pack, C.G, Kasai, R.S, Kubo, S, Schieber, N.L, Dohmae, N, Tochio, N, Hagiwara, K, Sasaki, Y, Aida, Y, Fujimori, F, Kigawa, T, Nishikori, K, Parton, R.G, Kusumi, A, Sako, Y, Anderluh, G, Yamashita, M, Kobayashi, T, Greimel, P, Kobayashi, T. | | Deposit date: | 2016-10-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C
FASEB J., 31, 2017
|
|
5H1A
 
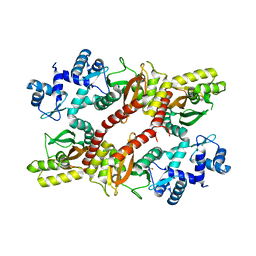 | | Crystal structure of an IclR homolog from Microbacterium sp. strain HM58-2 | | Descriptor: | IclR transcription factor homolog, PHOSPHATE ION | | Authors: | Akiyama, T, Yamada, Y, Takaya, N, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an IclR homologue from Microbacterium sp. strain HM58-2.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6KTK
 
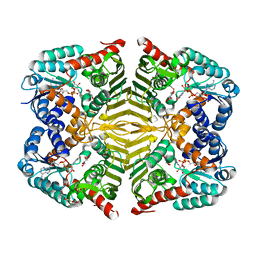 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NADH and L-glucono-1,5-lactone, from Paracoccus laeviglucosivorans | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTL
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTJ
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, apo-form, from Paracoccus laeviglucosivorans | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
5YAQ
 
 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with scyllo-inosose | | Descriptor: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YAB
 
 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YAP
 
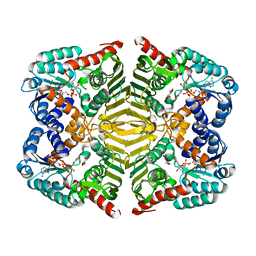 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with L-glucono-1,5-lactone | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YA8
 
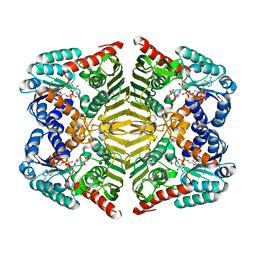 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
6IY9
 
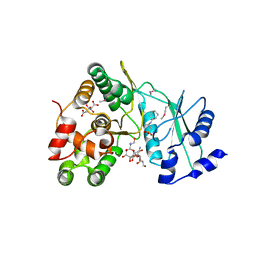 | | Crystal structure of aminoglycoside 7"-phoshotransferase-Ia (APH(7")-Ia/HYG) from Streptomyces hygroscopicus complexed with hygromycin B | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CITRATE ANION, HYGROMYCIN B VARIANT, ... | | Authors: | Takenoya, M, Shimamura, T, Yamanaka, R, Adachi, Y, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2018-12-14 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the substrate recognition of aminoglycoside 7''-phosphotransferase-Ia from Streptomyces hygroscopicus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
2EGH
 
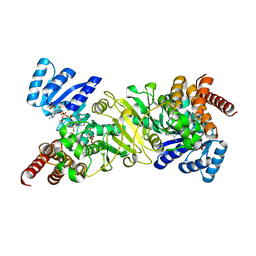 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase complexed with a magnesium ion, NADPH and fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, ... | | Authors: | Yajima, S, Hara, K, Iino, D, Sasaki, Y, Kuzuyama, T, Seto, H. | | Deposit date: | 2007-03-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in a quaternary complex with a magnesium ion, NADPH and the antimalarial drug fosmidomycin
Acta Crystallogr.,Sect.F, 63, 2007
|
|
7DQB
 
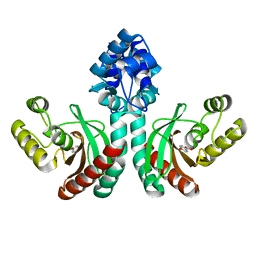 | | Crystal structure of an IclR homolog complexed with 4-hydroxybenzoate from Microbacterium hydrocarbonoxydans in P212121 form | | Descriptor: | IclR homolog, P-HYDROXYBENZOIC ACID | | Authors: | Akiyama, T, Sasaki, Y, Ito, S, Yajima, S. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis of the conformational changes in Microbacterium hydrocarbonoxydans IclR transcription factor homolog due to ligand binding.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
7CUO
 
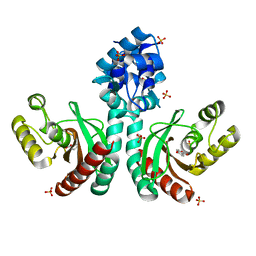 | | IclR transcription factor complexed with 4-hydroxybenzoic acid from Microbacterium hydrocarbonoxydans | | Descriptor: | P-HYDROXYBENZOIC ACID, SULFATE ION, Transcription factor | | Authors: | Akiyama, T, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-08-23 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the conformational changes in Microbacterium hydrocarbonoxydans IclR transcription factor homolog due to ligand binding.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
2RSY
 
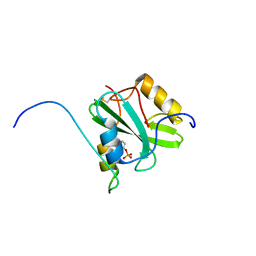 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
7D5N
 
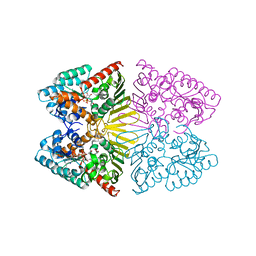 | | Crystal structure of inositol dehydrogenase homolog complexed with NADH and myo-inositol from Azotobacter vinelandii | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NADH and myo-inositol from Azotobacter vinelandii
To Be Published
|
|
7D5M
 
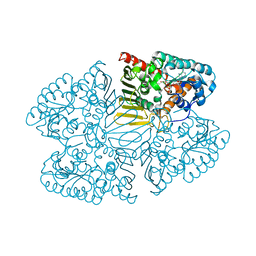 | | Crystal structure of inositol dehydrogenase homolog complexed with NAD+ from Azotobacter vinelandii | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase | | Authors: | Fukano, K, Ono, T, Suzuki, M, Takenoya, M, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of inositol dehydrogenase complexed with NAD+ from Azotobacter vinelandii
To Be Published
|
|
3A14
 
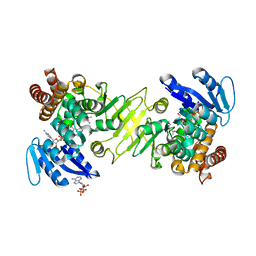 | | Crystal structure of DXR from Thermotoga maritima, in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MAGNESIUM ION, ... | | Authors: | Takenoya, M, Ohtaki, A, Noguchi, K, Sasaki, Y, Ohsawa, K, Yohda, M, Yajima, S. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase from the hyperthermophile Thermotoga maritima for insights into the coordination of conformational changes and an inhibitor binding.
J.Struct.Biol., 2010
|
|
3A06
 
 | | Crystal structure of DXR from Thermooga maritia, in complex with fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, ... | | Authors: | Takenoya, M, Ohtaki, A, Noguchi, K, Sasaki, Y, Ohsawa, K, Yohda, M, Yajima, S. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase from the hyperthermophile Thermotoga maritima for insights into the coordination of conformational changes and an inhibitor binding
J.Struct.Biol., 170, 2010
|
|
3W0R
 
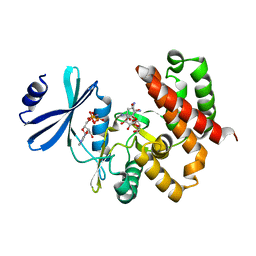 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (N202A), ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
3W0Q
 
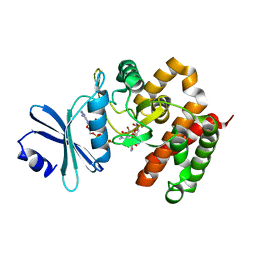 | | Crystal structure of a thermostable mutant of aminoglycoside phosphotransferase APH(4)-Ia (N203A), ternary complex with AMP-PNP and hygromycin B | | Descriptor: | HYGROMYCIN B VARIANT, Hygromycin-B 4-O-kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Iino, D, Takakura, Y, Fukano, K, Sasaki, Y, Hoshino, T, Ohsawa, K, Nakamura, A, Yajima, S. | | Deposit date: | 2012-11-02 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the ternary complex of APH(4)-Ia/Hph with hygromycin B and an ATP analog using a thermostable mutant.
J.Struct.Biol., 183, 2013
|
|
