7RVZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI26 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S71
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI35 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(hexylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6X
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI30 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S75
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI42 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(3-methylbutanoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S72
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI36 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6Y
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI32 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-[(cyclopropylmethyl)amino]-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S73
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI37 | | Descriptor: | (6S)-5-{(2S)-2-[(tert-butylcarbamoyl)amino]-3,3-dimethylbutanoyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-5-azaspiro[2.4]heptane-6-carboxamide (non-preferred name), 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6Z
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S70
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI34 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(butylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
5TPH
 
 | | Crystal structure of a de novo designed protein homodimer with curved beta-sheet | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, de novo NTF2 homodimer | | Authors: | Basanta, B, Marcos, E, Oberdorfer, G, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TS4
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Chidyausiku, T.M, Marcos, E, Pereira, J.H, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
4E6T
 
 | | Structure of LpxA from Acinetobacter baumannii at 1.8A resolution (P212121 form) | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CITRATE ANION | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2012-03-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of LpxA from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3NMD
 
 | | Crystal structure of the leucine zipper domain of cGMP dependent protein kinase I beta | | Descriptor: | GLYCEROL, HEXANE-1,6-DIOL, cGMP Dependent PRotein Kinase | | Authors: | Kim, C, Casteel, D.E, Smith-Nguyen, E.V, Sankaran, B, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2010-06-22 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | A crystal structure of the cyclic GMP-dependent protein kinase I{beta} dimerization/docking domain reveals molecular details of isoform-specific anchoring.
J.Biol.Chem., 285, 2010
|
|
7RVO
 
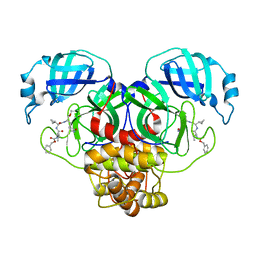 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI13 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW1
 
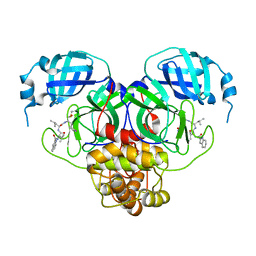 | |
7RVP
 
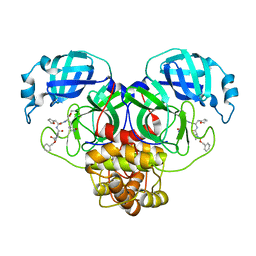 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI14 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-furan-2-yl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVT
 
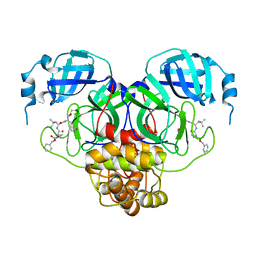 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI20 | | Descriptor: | 3C-like proteinase, N~2~-[(2S)-2-{[(benzyloxy)carbonyl]amino}-2-cyclopropylacetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6W
 
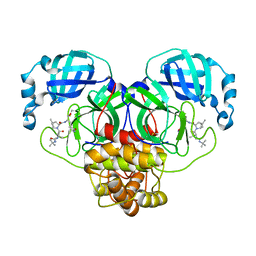 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI29 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S74
 
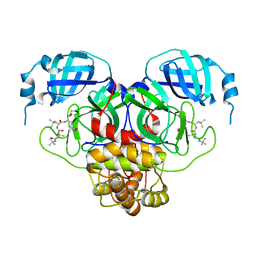 | |
9D9T
 
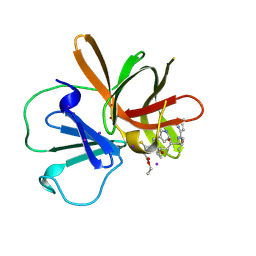 | | Human norovirus GII.4 Sydney protease R112A mutant in complex with rupintrivir | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, IODIDE ION, Peptidase C37 | | Authors: | Pham, S.H, Neetu, N, Sankaran, B, Prasad, B.V.V. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational flexibility is a critical factor in designing broad-spectrum human norovirus protease inhibitors.
J.Virol., 99, 2025
|
|
5C6C
 
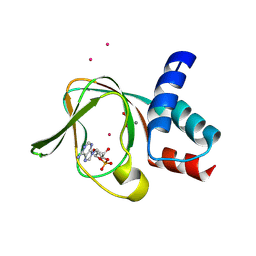 | | PKG II's Amino Terminal Cyclic Nucleotide Binding Domain (CNB-A) in a complex with cAMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CADMIUM ION, ... | | Authors: | Campbell, J.C, Reger, A.S, Huang, G.Y, Sankaran, B, Kim, J.J, Kim, C.W. | | Deposit date: | 2015-06-22 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Cyclic Nucleotide Selectivity in cGMP-dependent Protein Kinase II.
J.Biol.Chem., 291, 2016
|
|
5TVV
 
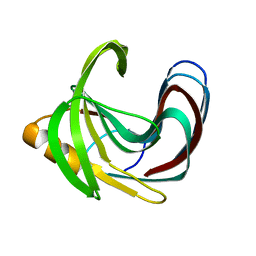 | | Computationally Designed Fentanyl Binder - Fen49* Apo | | Descriptor: | Endo-1,4-beta-xylanase A, POTASSIUM ION | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, A.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-10 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
9CKX
 
 | |
5U35
 
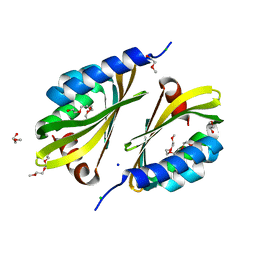 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
3MYU
 
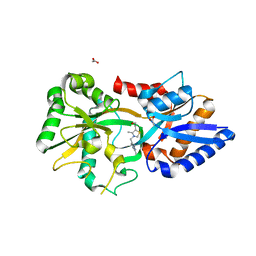 | | Mycoplasma genitalium MG289 | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ACETATE ION, High affinity transport system protein p37 | | Authors: | Sippel, K.H, Boehlein, S.K, Govindasamy, L, Namiki, K, Satai, Y, Quirit, J.G, Agbandje-McKenna, M, Goodison, S, Rosser, C.J, Sankaran, B, McKenna, R. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into Mycoplasma genitalium metabolism revealed by the structure of MG289, an extracytoplasmic thiamine binding lipoprotein.
Proteins, 79, 2011
|
|
