7MP0
 
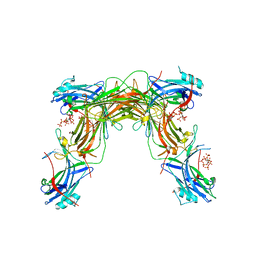 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 5-DIPHOSPHATE PENTAKISPHOSPHATE (5-PP IP5) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, 2-ETHOXYETHANOL, S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7MOR
 
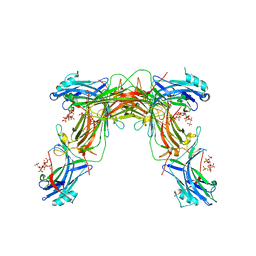 | |
7MP2
 
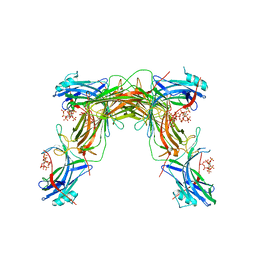 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1D-MYO-INOSITOL 1,5-BISDIPHOSPHATE TETRAKISPHOSPHATE (1,5-PP IP4) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], S-arrestin | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7MP1
 
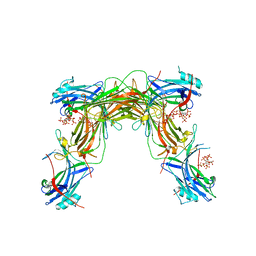 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH 1,5-DI-METHYLENEBISPHOSPHONATE INOSITOL TETRAKISPHOSPHATE (1,5-PCP-IP4) | | Descriptor: | S-arrestin, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(hydroxyphosphoryl)methanediyl]}bis(phosphonic acid) | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2021-05-04 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7JXA
 
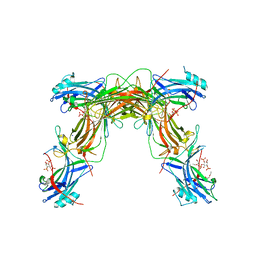 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH INOSITOL 1,4,5-TRIPHOSPHATE | | Descriptor: | 2-ETHOXYETHANOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, S-arrestin, ... | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7JSM
 
 | |
7JTB
 
 | |
2V9P
 
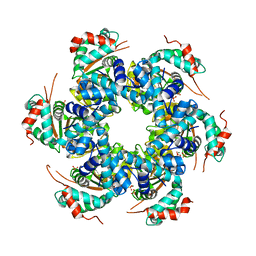 | | Crystal structure of papillomavirus E1 hexameric helicase DNA-free form | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, REPLICATION PROTEIN E1 | | Authors: | Sanders, C.M, Kovalevskiy, O.V, Sizov, D, Lebedev, A.A, Isupov, M.N, Antson, A.A. | | Deposit date: | 2007-08-24 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Papillomavirus E1 Helicase Assembly Maintains an Asymmetric State in the Absence of DNA and Nucleotide Cofactors.
Nucleic Acids Res., 35, 2007
|
|
2JEX
 
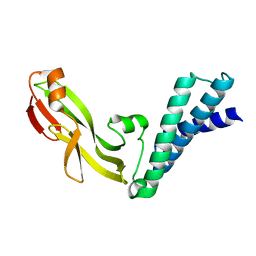 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-24 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
2JEU
 
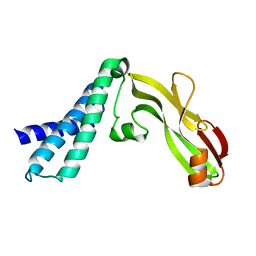 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-23 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
2NDJ
 
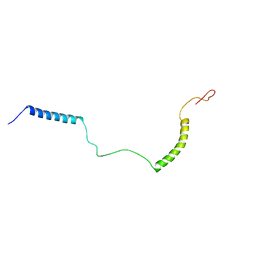 | | Structural Basis for KCNE3 and Estrogen Modulation of the KCNQ1 Channel | | Descriptor: | Potassium voltage-gated channel subfamily E member 3 | | Authors: | Sanders, C.R, Van Horn, W.D, Kroncke, B.M, Sisco, N.J, Meiler, J, Vanoye, C.G, Song, Y, Nannemann, D.P, Welch, R.C, Kang, C, Smith, J, George, A.L. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for KCNE3 modulation of potassium recycling in epithelia.
Sci Adv, 2, 2016
|
|
8GII
 
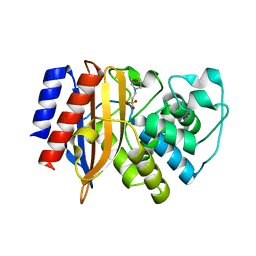 | | TEM-1 Beta Lactamase Variant 80.a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TEM-1 Variant 80.a | | Authors: | Fram, B.F, Gauthier, N.P, Khan, A.R, Sander, C. | | Deposit date: | 2023-03-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Simultaneous enhancement of multiple functional properties using evolution-informed protein design.
Nat Commun, 15, 2024
|
|
8GIJ
 
 | |
8RQU
 
 | | Structure of TEM1 beta-lactamase variant 70.a | | Descriptor: | Beta-lactamase TEM-1, MAGNESIUM ION | | Authors: | Napier, E, Fram, B.F, Gauthier, N.P, Sander, C, Khan, A.R. | | Deposit date: | 2024-01-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Simultaneous enhancement of multiple functional properties using evolution-informed protein design.
Nat Commun, 15, 2024
|
|
1RPR
 
 | | THE STRUCTURE OF COLE1 ROP IN SOLUTION | | Descriptor: | ROP | | Authors: | Eberle, W, Pastore, A, Klaus, W, Sander, C, Roesch, P. | | Deposit date: | 1991-10-09 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of ColE1 rop in solution.
J.Biomol.NMR, 1, 1991
|
|
2N44
 
 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N45
 
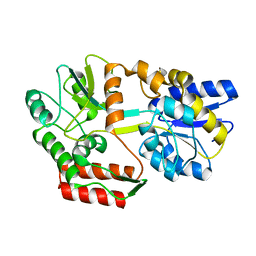 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data with a second set of RDC data simulated for an alternative alignment tensor. Northeast Structural Genomics Consortium target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N42
 
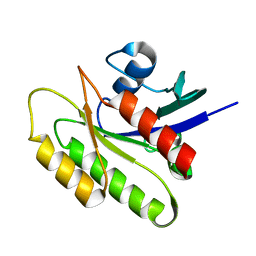 | | EC-NMR Structure of Human H-RasT35S mutant protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data | | Descriptor: | GTPase HRas | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N47
 
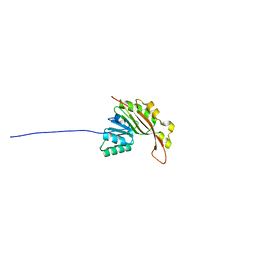 | | EC-NMR Structure of Synechocystis sp. PCC 6803 Slr1183 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N4F
 
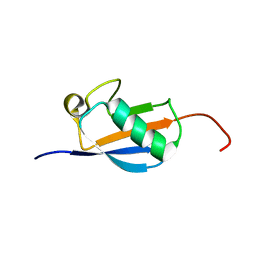 | | EC-NMR Structure of Arabidopsis thaliana At2g32350 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AR3433A | | Descriptor: | uncharacterized protein AR3433A | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N4A
 
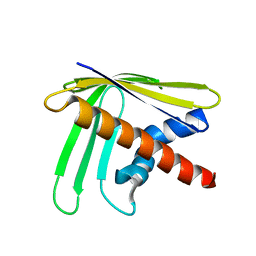 | | EC-NMR Structure of Ralstonia metallidurans Rmet_5065 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target CrR115 | | Descriptor: | Uncharacterized protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N49
 
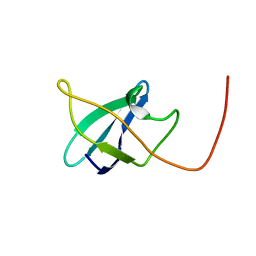 | | EC-NMR Structure of Erwinia carotovora ECA1580 N-terminal Domain Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target EwR156A | | Descriptor: | Putative cold-shock protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N4D
 
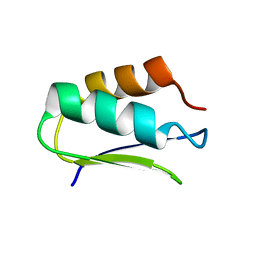 | | EC-NMR Structure of Agrobacterium tumefaciens Atu1203 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target AtT10 | | Descriptor: | Uncharacterized protein Atu1203 | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N46
 
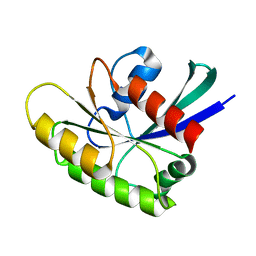 | | EC-NMR Structure of Human H-RasT35S mutant protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data | | Descriptor: | GTPase HRas | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
2N4B
 
 | | EC-NMR Structure of Ralstonia metallidurans Rmet_5065 Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target CrR115 | | Descriptor: | Uncharacterized protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
