4KPR
 
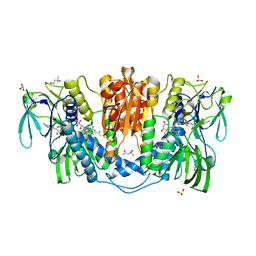 | | Tetrameric form of rat selenoprotein thioredoxin reductase 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, SULFITE ION, ... | | Authors: | Lindqvist, Y, Sandalova, T, Xu, J, Arner, E. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
To be Published, 2013
|
|
5M00
 
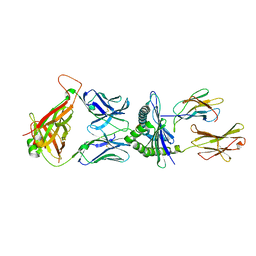 | | Crystal structure of murine P14 TCR complex with H-2Db and Y4A, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
3TBS
 
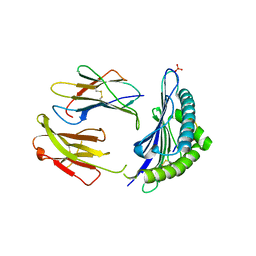 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX THE WITH LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (V3P,Y4A) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCOPROTEIN G1, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
3TBV
 
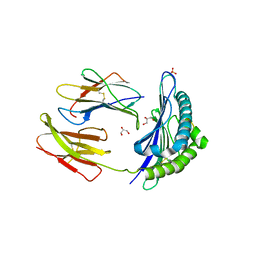 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (A2G,V3P,Y4A) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Glycoprotein G1, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
3TBY
 
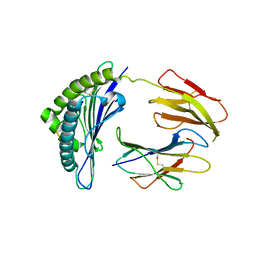 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (V3P, Y4F) | | Descriptor: | Beta-2-microglobulin, GLYCOPROTEIN GPC, H-2 class I histocompatibility antigen, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
5CTV
 
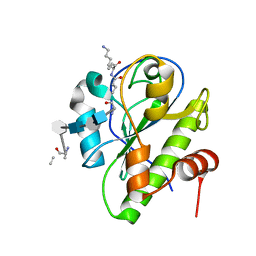 | | Catalytic domain of LytA, the major autolysin of Streptococcus pneumoniae, (C60A, H133A, C136A mutant) complexed with peptidoglycan fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, Autolysin, fragment of peptidoglycan | | Authors: | Achour, A, Sandalova, T, Mellroth, P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Mol.Microbiol., 101, 2016
|
|
1E3L
 
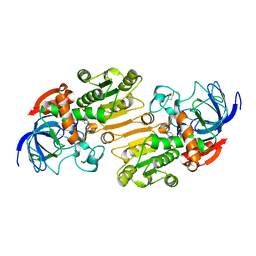 | | P47H mutant of mouse class II alcohol dehydrogenase complex with NADH | | Descriptor: | ALCOHOL DEHYDROGENASE, CLASS II, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Svensson, S, Hoog, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-19 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E3I
 
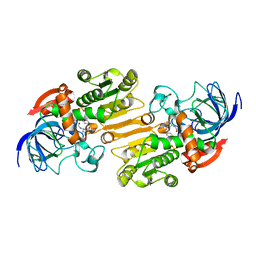 | | Mouse class II alcohol dehydrogenase complex with NADH and inhibitor | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, CLASS II, ... | | Authors: | Svensson, S, Hoog, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-16 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
1E3E
 
 | | Mouse class II alcohol dehydrogenase complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, CLASS II, ... | | Authors: | Svensson, S, Hoeoeg, J.O, Schneider, G, Sandalova, T. | | Deposit date: | 2000-06-14 | | Release date: | 2000-09-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of Mouse Class II Alcohol Dehydrogenase Reveal Determinants of Substrate Specificity and Catalytic Efficiency
J.Mol.Biol., 302, 2000
|
|
3TBT
 
 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (V3P, Y4S) | | Descriptor: | Beta-2-microglobulin, GLYCOPROTEIN G1, H-2 class I histocompatibility antigen, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
1OVM
 
 | | Crystal structure of Indolepyruvate decarboxylase from Enterobacter cloacae | | Descriptor: | Indole-3-pyruvate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Schutz, A, Sandalova, T, Ricagno, S, Hubner, G, Konig, S, Schneider, G. | | Deposit date: | 2003-03-27 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of thiamindiphosphate-dependent indolepyruvate decarboxylase from Enterobacter cloacae, an enzyme involved in the biosynthesis of the plant hormone indole-3-acetic acid
Eur.J.Biochem., 270, 2003
|
|
3TBW
 
 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (A2G, V3P, Y4S) | | Descriptor: | Beta-2-microglobulin, GLYCOPROTEIN GPC, H-2 class I histocompatibility antigen, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
4STD
 
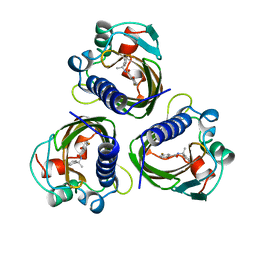 | | HIGH RESOLUTION STRUCTURES OF SCYTALONE DEHYDRATASE-INHIBITOR COMPLEXES CRYSTALLIZED AT PHYSIOLOGICAL PH | | Descriptor: | N-[1-(4-BROMOPHENYL)ETHYL]-5-FLUORO SALICYLAMIDE, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
4S1F
 
 | | Fructose-6-phosphate aldolase A from E.coli soaked in acetylacetone | | Descriptor: | Fructose-6-phosphate aldolase 1, pentane-2,4-dione | | Authors: | Stellmacher, L, Sandalova, T, Leptihn, S, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Acid Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase
ChemCatChem, 2015
|
|
6G9Q
 
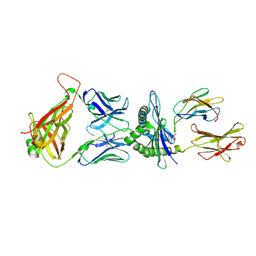 | |
6H6D
 
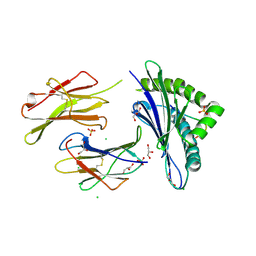 | |
6H6H
 
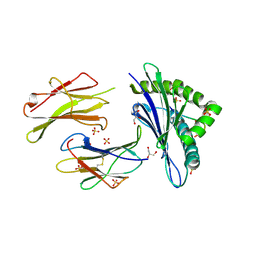 | |
1ZJ8
 
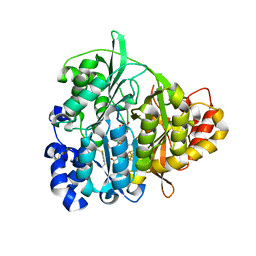 | | Structure of Mycobacterium tuberculosis NirA protein | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Probable ferredoxin-dependent nitrite reductase NirA, ... | | Authors: | Schnell, R, Sandalova, T, Hellman, U, Lindqvist, Y, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis Is a Sulfite Reductase with a Covalent Cys-Tyr Bond in the Active Site
J.Biol.Chem., 280, 2005
|
|
1ZJ9
 
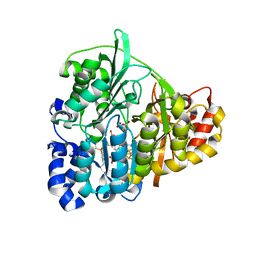 | | Structure of Mycobacterium tuberculosis NirA protein | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Probable ferredoxin-dependent nitrite reductase NirA, ... | | Authors: | Schnell, R, Sandalova, T, Hellman, U, Lindqvist, Y, Schneider, G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis Is a Sulfite Reductase with a Covalent Cys-Tyr Bond in the Active Site
J.Biol.Chem., 280, 2005
|
|
4RXG
 
 | | Fructose-6-phosphate aldolase Q59E from E.coli | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL, PENTAETHYLENE GLYCOL | | Authors: | Stellmacher, L, Sandalova, T, Leptihn, S, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Acid Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase
ChemCatChem, 2015
|
|
4RXF
 
 | | Fructose-6-phosphate aldolase Y131F from E.coli | | Descriptor: | Fructose-6-phosphate aldolase 1, PHOSPHATE ION | | Authors: | Stellmacher, L, Sandalova, T, Leptihn, S, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Acid Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase
ChemCatChem, 2015
|
|
6G9R
 
 | |
6GB5
 
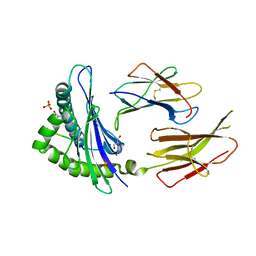 | | Structure of H-2Db with truncated SEV peptide and GL | | Descriptor: | Beta-2-microglobulin, GLY-LEU, GLYCEROL, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GB7
 
 | | Structure of H-2Db with scoop loop from tapasin | | Descriptor: | Beta-2-microglobulin, GLY-GLY-LEU-SER, GLYCEROL, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4EX6
 
 | | Crystal structure of the alnumycin P phosphatase AlnB | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
