1XWU
 
 | | Solution structure of ACAUAGA loop | | Descriptor: | 5'-R(*CP*GP*AP*AP*AP*CP*AP*UP*AP*GP*AP*UP*UP*CP*GP*A)-3' | | Authors: | Sakamoto, T, Oguro, A, Kawai, G, Ohtsu, T, Nakamura, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structures of double loops of an RNA aptamer against mammalian initiation factor 4A
Nucleic Acids Res., 33, 2005
|
|
1XWP
 
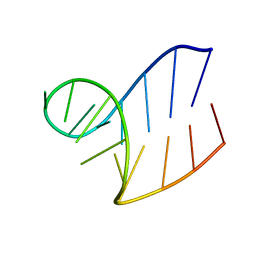 | | Solution structure of AUCGCA loop | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*UP*CP*GP*CP*AP*CP*UP*CP*CP*A)-3' | | Authors: | Sakamoto, T, Oguro, A, Kawai, G, Ohtsu, T, Nakamura, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structures of double loops of an RNA aptamer against mammalian initiation factor 4A
Nucleic Acids Res., 33, 2005
|
|
1L1W
 
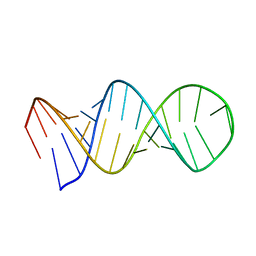 | | NMR structure of a SRP19 binding domain in human SRP RNA | | Descriptor: | SRP19 binding domain of SRP RNA | | Authors: | Sakamoto, T, Morita, S, Tabata, K, Nakamura, K, Kawai, G. | | Deposit date: | 2002-02-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a SRP19 binding domain in human SRP RNA.
J.Biochem.(Tokyo), 132, 2002
|
|
7YQS
 
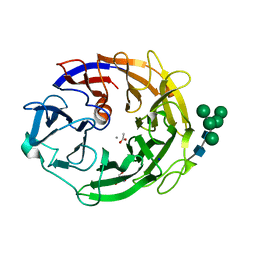 | | Neutron structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.25 Å), X-RAY DIFFRACTION | | Cite: | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
2RRC
 
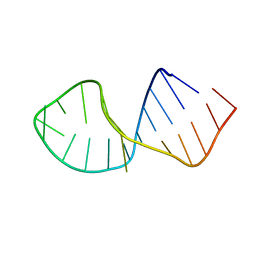 | | Solution Structure of RNA aptamer against AML1 Runt domain | | Descriptor: | 5'-R(P*GP*GP*AP*CP*CP*CP*(AP7)P*CP*CP*AP*CP*GP*GP*CP*GP*AP*GP*GP*UP*CP*CP*A)-3' | | Authors: | Nomura, Y, Fujiwara, K, Chiba, M, Fukunaga, J, Tanaka, Y, Iibuchi, H, Tanaka, T, Nakamura, Y, Kawai, G, Kozu, T, Sakamoto, T. | | Deposit date: | 2010-06-23 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A novel high affinity RNA motif that mimics DNA in AML1 Runt domain binding
To be Published
|
|
8WO8
 
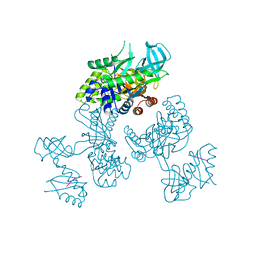 | | Crystal Structure of an RNA-binding protein, FAU-1, from Pyrococcus furiosus | | Descriptor: | Probable ribonuclease FAU-1, RNA (5'-R(P*AP*UP*A)-3') | | Authors: | Kawai, G, Okada, K, Baba, S, Sato, A, Sakamoto, T, Kanai, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Homo-trimeric structure of the ribonuclease for rRNA processing, FAU-1, from Pyrococcus furiosus.
J.Biochem., 175, 2024
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
7VH9
 
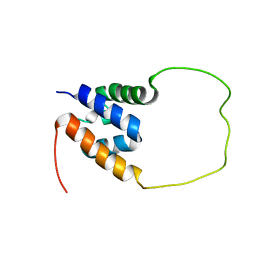 | | Solution structure of the chimeric peptide of the first SURP domain of Human SF3A1 and the interacting region of SF1. | | Descriptor: | Splicing factor 3A subunit 1,Splicing factor 1 | | Authors: | Muto, Y, Kuwasako, K, Takizawa, M, Kobayashi, N, Sakamoto, T. | | Deposit date: | 2021-09-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction between the first SURP domain of the SF3A1 subunit in U2 snRNP and the human splicing factor SF1.
Protein Sci., 31, 2022
|
|
8I4D
 
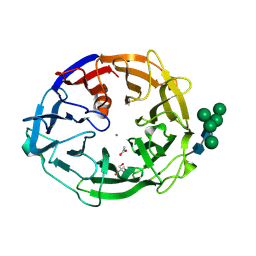 | | X-ray structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
8IHW
 
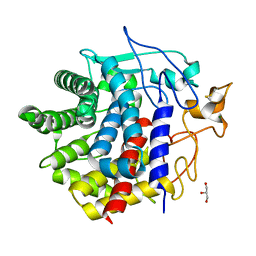 | | X-ray crystal structure of D43R mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHX
 
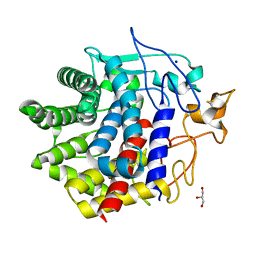 | | X-ray crystal structure of N372D mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHY
 
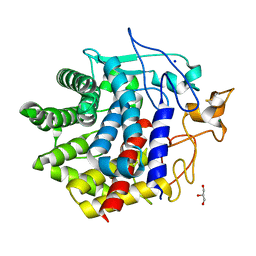 | | X-ray crystal structure of Q387E mutant of endo-1,4-beta glucanase from Eisenia fetida | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
2FYH
 
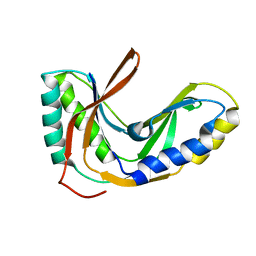 | | Solution structure of the 2'-5' RNA ligase-like protein from Pyrococcus furiosus | | Descriptor: | putative integral membrane transport protein | | Authors: | Okada, K, Matsuda, T, Sakamoto, T, Muto, Y, Yokoyama, S, Kanai, A, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of a heat-stable enzyme possessing GTP-dependent RNA ligase activity from a hyperthermophilic archaeon, Pyrococcus furiosus
Rna, 15, 2009
|
|
6IIE
 
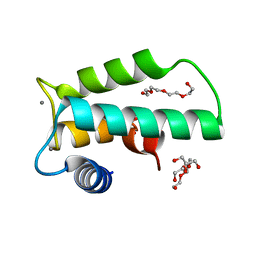 | | Crystal structure of human diacylglycerol kinase alpha EF-hand domains bound to Ca2+ | | Descriptor: | CALCIUM ION, Diacylglycerol kinase alpha, GLYCEROL, ... | | Authors: | Takahashi, D, Suzuki, K, Sakamoto, T, Iwamoto, T, Murata, T, Sakane, F. | | Deposit date: | 2018-10-04 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Crystal structure and calcium-induced conformational changes of diacylglycerol kinase alpha EF-hand domains.
Protein Sci., 28, 2019
|
|
2F87
 
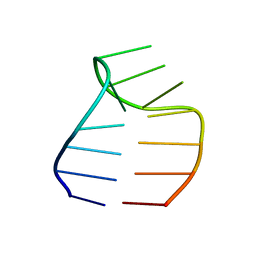 | | Solution structure of a GAAG tetraloop in SRP RNA from Pyrococcus furiosus | | Descriptor: | SRP RNA | | Authors: | Okada, K, Takahashi, M, Sakamoto, T, Nakamura, K, Kanai, A, Kawai, G | | Deposit date: | 2005-12-02 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a GAAG tetraloop in helix 6 of SRP RNA from Pyrococcus furiosus
Nucleosides Nucleotides Nucleic Acids, 25, 2006
|
|
7DFS
 
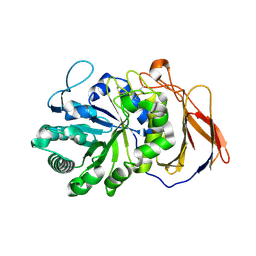 | | Crystal structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S - Rha-GlcA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase, alpha-D-mannopyranose, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7DFQ
 
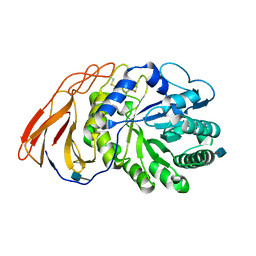 | | Crystal Structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7ESK
 
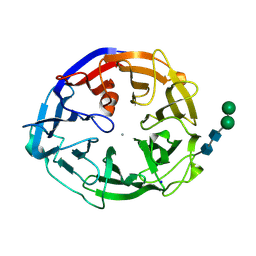 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, Ligand free form | | Descriptor: | CALCIUM ION, L-Rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESM
 
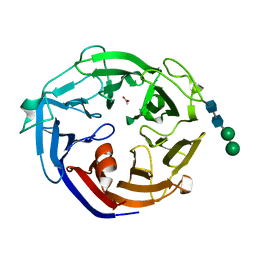 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | Descriptor: | ACETATE ION, L-rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESN
 
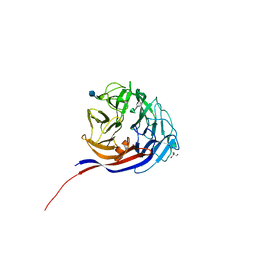 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, H105F Rha-GlcA complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, L-Rhamnose-alpha-1,4-D-glucuronate lyase, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESL
 
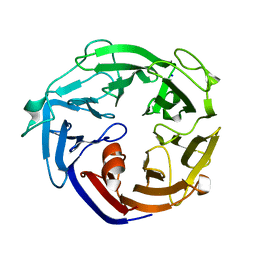 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, N247A N-glycan free form | | Descriptor: | L-rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
5XQJ
 
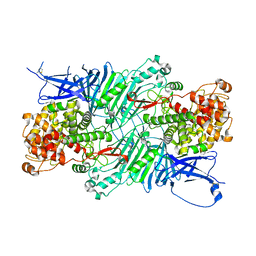 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
5XQG
 
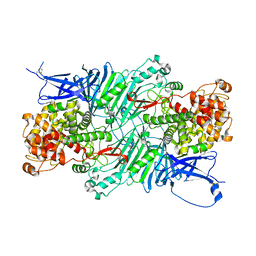 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, Pcrglx protein | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
5XQ3
 
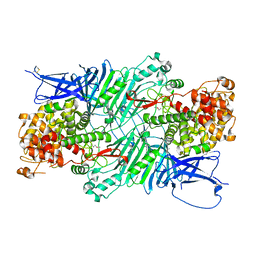 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum | | Descriptor: | CALCIUM ION, Pcrglx protein | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-06 | | Release date: | 2018-03-21 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
5XQO
 
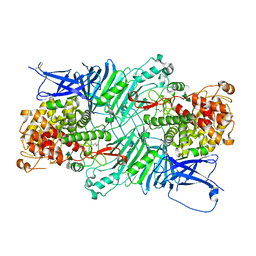 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with tetrameric substrate | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose-(1-4)-alpha-D-galactopyranuronic acid-(1-2)-alpha-L-rhamnopyranose, 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-alpha-L-rhamnopyranose-(1-4)-alpha-D-galactopyranuronic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
