1FYW
 
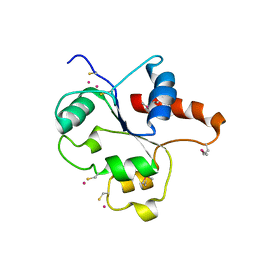 | | CRYSTAL STRUCTURE OF THE TIR DOMAIN OF HUMAN TLR2 | | Descriptor: | TOLL-LIKE RECEPTOR 2 | | Authors: | Xu, Y, Tao, X, Shen, B, Horng, T, Medzhitov, R, Manley, J.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for signal transduction by the Toll/interleukin-1 receptor domains.
Nature, 408, 2000
|
|
2CAN
 
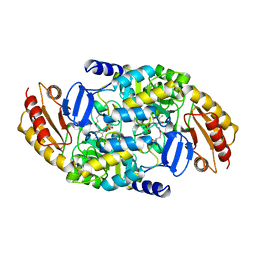 | |
7N7V
 
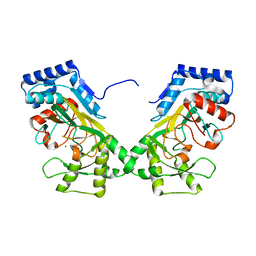 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
6XE0
 
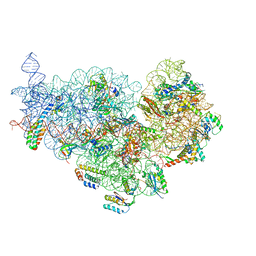 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4HZP
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, CHLORIDE ION, COENZYME A, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
4HZN
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional Methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, GLYCEROL, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
4HZO
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP acyltransferase/decarboxylase, CHLORIDE ION, COENZYME A | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
8G5T
 
 | | Crystal structure of apo TnmK2 | | Descriptor: | TnmK2 | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2023-02-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis.
Nat.Chem.Biol., 20, 2024
|
|
8G5S
 
 | | Crystal structure of apo TnmJ | | Descriptor: | TnmJ | | Authors: | Liu, Y.-C, Li, G, Gui, C, Shen, B. | | Deposit date: | 2023-02-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis.
Nat.Chem.Biol., 20, 2024
|
|
8G5U
 
 | | Crystal structure of TnmK2 complexed with TNM B | | Descriptor: | TnmK2, methyl (2E)-3-[(1aS,11S,11aS,14Z,18R)-3,18-dihydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]but-2-enoate | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2023-02-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis.
Nat.Chem.Biol., 20, 2024
|
|
1GBN
 
 | | HUMAN ORNITHINE AMINOTRANSFERASE COMPLEXED WITH THE NEUROTOXIN GABACULINE | | Descriptor: | 3-AMINOBENZOIC ACID, GABACULINE, ORNITHINE AMINOTRANSFERASE, ... | | Authors: | Shah, S.A, Shen, B.W, Brunger, A.T. | | Deposit date: | 1997-05-29 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human ornithine aminotransferase complexed with L-canaline and gabaculine: structural basis for substrate recognition.
Structure, 5, 1997
|
|
9CEN
 
 | |
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
4Z5Q
 
 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
6DA9
 
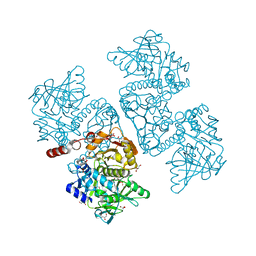 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | Authors: | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-12-06 | | Release date: | 2013-02-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
1D7P
 
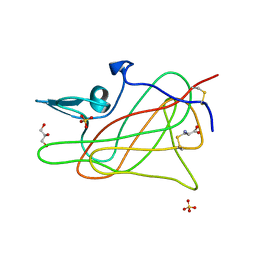 | | Crystal structure of the c2 domain of human factor viii at 1.5 a resolution at 1.5 A | | Descriptor: | COAGULATION FACTOR VIII PRECURSOR, CYSTEINE, GLYCEROL, ... | | Authors: | Pratt, K.P, Shen, B.W, Stoddard, B.L. | | Deposit date: | 1999-10-19 | | Release date: | 1999-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the C2 domain of human factor VIII at 1.5 A resolution.
Nature, 402, 1999
|
|
4ZDN
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-17 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6OMQ
 
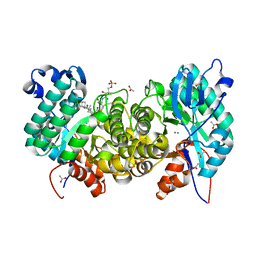 | | Crystal structure of PtmU3 complexed with PTM substrate | | Descriptor: | ACETATE ION, MANGANESE (II) ION, PtmU3, ... | | Authors: | Liu, Y.C, Dong, L.B, Shen, B. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterization and Crystal Structure of a Nonheme Diiron Monooxygenase Involved in Platensimycin and Platencin Biosynthesis.
J.Am.Chem.Soc., 141, 2019
|
|
5BMO
 
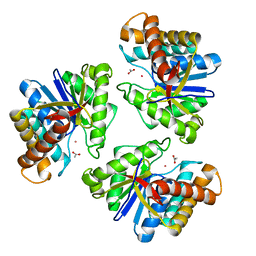 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
1Q1C
 
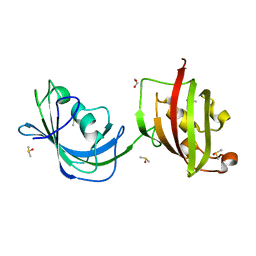 | | Crystal structure of N(1-260) of human FKBP52 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-07-18 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5BP8
 
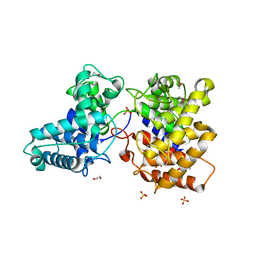 | | ent-Copalyl diphosphate synthase from Streptomyces platensis | | Descriptor: | 1,2-ETHANEDIOL, Ent-copalyl diphosphate synthase, SULFATE ION | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Ma, M, Chang, C.-Y, Shen, B, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the ent-Copalyl Diphosphate Synthase PtmT2 from Streptomyces platensis CB00739, a Bacterial Type II Diterpene Synthase.
J.Am.Chem.Soc., 138, 2016
|
|
1N1A
 
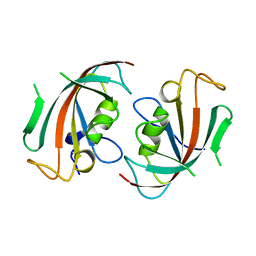 | | Crystal Structure of the N-terminal domain of human FKBP52 | | Descriptor: | FKBP52 | | Authors: | Li, P, Ding, Y, Wu, B, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the N-terminal domain of human FKBP52.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
