3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
3CNG
 
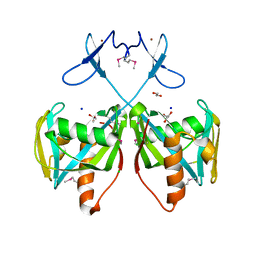 | | Crystal structure of NUDIX hydrolase from Nitrosomonas europaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
3OT1
 
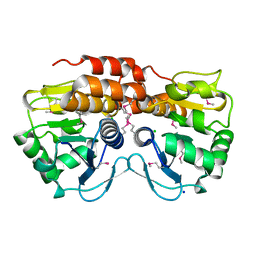 | | Crystal structure of VC2308 protein | | Descriptor: | 4-methyl-5(B-hydroxyethyl)-thiazole monophosphate biosynthesis enzyme, CHLORIDE ION, SODIUM ION | | Authors: | Niedzialkowska, E, Wawrzak, Z, Chruszcz, M, Porebski, P, Skarina, T, Huang, X, Grimshaw, S, Cymborowski, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-10 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of VC2308 protein
To be Published
|
|
4PVA
 
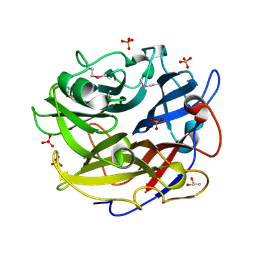 | | Crystal structure of GH62 hydrolase from thermophilic fungus Scytalidium thermophilum | | Descriptor: | GH62 hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Nocek, B, Kaur, A.P, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2014-03-15 | | Release date: | 2014-11-19 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Functional and structural diversity in GH62 alpha-L-arabinofuranosidases from the thermophilic fungus Scytalidium thermophilum.
Microb Biotechnol, 8, 2015
|
|
3GC2
 
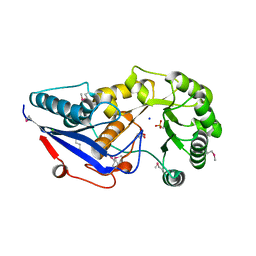 | | 1.85 Angstrom Crystal Structure of O-succinylbenzoate Synthase from Salmonella typhimurium in Complex with Succinic Acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, SODIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-21 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of O-succinylbenzoate Synthase
from Salmonella typhimurium in Complex with Succinic Acid
To be Published
|
|
5HNM
 
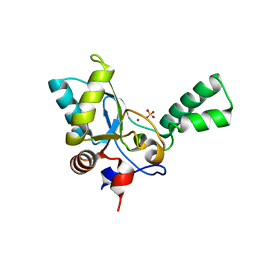 | | Crystal structure of vancomycin resistance D,D-pentapeptidase VanY E175A mutant from VanB-type resistance cassette in complex with Zn(II) | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, SULFATE ION, ZINC ION | | Authors: | Stogios, P.J, Chun, J, Wawrzak, Z, Evdokimova, E, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | To be published
To Be Published
|
|
3GE1
 
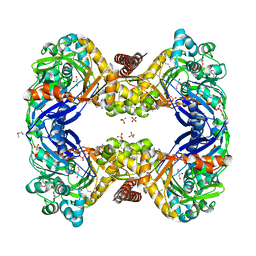 | | 2.7 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with ADP and Glycerol | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Brunzelle, J, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with ADP and Glycerol
To be Published
|
|
3DCL
 
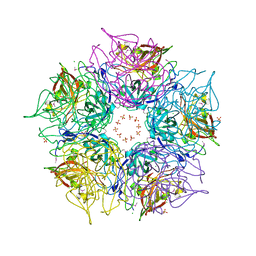 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chruszcz, M, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
3DCA
 
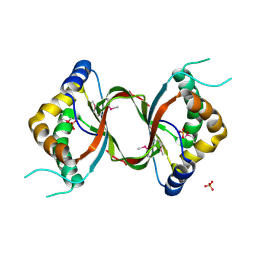 | | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target | | Descriptor: | RPA0582, SULFATE ION | | Authors: | Sledz, P, Wang, S, Chruszcz, M, Yim, V, Kudritska, M, Evdokimova, E, Turk, D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-05 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target
To be Published
|
|
4PSU
 
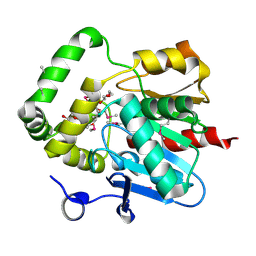 | | Crystal structure of alpha/beta hydrolase from Rhodopseudomonas palustris CGA009 | | Descriptor: | Alpha/beta hydrolase, DODECAETHYLENE GLYCOL | | Authors: | Nocek, B, Hajighasemi, M, Xu, X, Cui, H, Savchenko, A, Yakunin, A. | | Deposit date: | 2014-03-07 | | Release date: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of alpha/beta hydrolase from Rhodopseudomonas palustris CGA009
TO BE PUBLISHED
|
|
3DR6
 
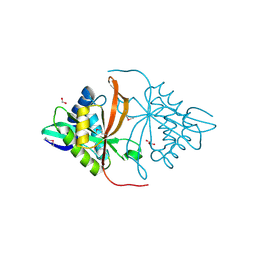 | | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, yncA | | Authors: | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-10 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Funded by the national institute of
allergy and infectious diseases of nih (contract number
hhsn272200700058c).
To be Published
|
|
6O6Q
 
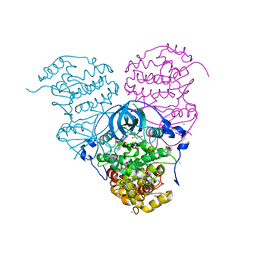 | | Crystal structure of Cka1p, a casein kinase 2 alpha ortholog from Candida albicans | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Casein kinase 2 catalytic subunit, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Cka1p, a casein kinase 2 alpha ortholog from Candida albicans
To Be Published
|
|
1SH8
 
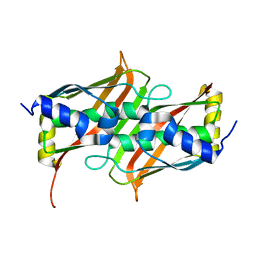 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA5026 from Pseudomonas aeruginosa, Probable Thioesterase | | Descriptor: | hypothetical protein PA5026 | | Authors: | Zhang, R, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA5026 from Pseudomonas aeruginosa
To be Published
|
|
1SED
 
 | | Crystal Structure of Protein of Unknown Function YhaL from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Hypothetical protein yhaI, ... | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Hypothetical Protein YhaI, APC1180 from Bacillus subtilis
To be Published
|
|
3GEM
 
 | | Crystal structure of short-chain dehydrogenase from Pseudomonas syringae | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Nocek, B, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of short-chain dehydrogenase from Pseudomonas syringae.
To be Published
|
|
3GHJ
 
 | | Crystal structure from the mobile metagenome of Halifax Harbour Sewage Outfall: Integron Cassette Protein HFX_CASS4 | | Descriptor: | Putative integron gene cassette protein | | Authors: | Sureshan, V, Deshpande, C, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Kim, Y, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Structure from the mobile metagenome of Halifax Harbour Sewage Outfall: Integron Cassette Protein HFX_CASS4
To be Published
|
|
3DR3
 
 | | Structure of IDP00107, a potential N-acetyl-gamma-glutamylphosphate reductase from Shigella flexneri | | Descriptor: | D-MALATE, N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-10 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of IDP00107, a potential N-acetyl-gamma-glutamylphosphate reductase from Shigella flexneri
To be Published
|
|
1ORU
 
 | | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis | | Descriptor: | CHLORIDE ION, SULFATE ION, yuaD protein | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-15 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis
To be Published
|
|
1OTK
 
 | | Structural Genomics, Protein paaC | | Descriptor: | Phenylacetic acid degradation protein paaC | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-21 | | Release date: | 2003-10-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2 A crystal structure of protein paaC from E. Coli
To be Published
|
|
1OU0
 
 | | precorrin-8X methylmutase related protein | | Descriptor: | precorrin-8X methylmutase related protein | | Authors: | Cuff, M.E, Joachimiak, A, Korolev, S, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-24 | | Release date: | 2003-10-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a predicted precorrin-8x methylmutase from Thermoplasma acidophilum.
Proteins, 58, 2004
|
|
3E4F
 
 | | Crystal structure of BA2930- a putative aminoglycoside N3-acetyltransferase from Bacillus anthracis | | Descriptor: | Aminoglycoside N3-acetyltransferase, CITRIC ACID | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3E6Q
 
 | | Putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-15 | | Release date: | 2008-08-26 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure of putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa.
To be Published
|
|
3E9Q
 
 | | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-23 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Short Chain Dehydrogenase from Shigella flexneri
To be Published
|
|
3EET
 
 | | Crystal structure of putative GntR-family transcriptional regulator | | Descriptor: | Putative GntR-family transcriptional regulator | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-05 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Crystal structure of putative GntR-family transcriptional regulator from Streptomyces avermitilis
To be Published
|
|
3IMF
 
 | | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Gordon, E, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
