2BY8
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BYE
 
 | | NMR solution structure of phospholipase c epsilon RA 1 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2BYF
 
 | | NMR solution structure of phospholipase c epsilon RA 2 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2AP6
 
 | | X-Ray Crystal Structure of Protein Atu4242 from Agrobacterium tumefaciens. Northeast Strucutral Genomics Consortium Target AtR43. | | Descriptor: | hypothetical protein Atu4242 | | Authors: | Benach, J, Kuzin, A.P, Forouhar, F, Abashidze, M, Vorobiev, S.M, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a Hypothetical protein Atu4242 from Agrobacterium tumefaciens (strain C58 / ATCC 3 NESG Target ATR43.
To be Published
|
|
2ARA
 
 | |
1OZY
 
 | |
1SBK
 
 | | X-RAY STRUCTURE OF YDII_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER29. | | Descriptor: | Hypothetical protein ydiI, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Vorobiev, S.M, Lee, I, Forouhar, F, Ma, L, Chiang, Y, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Structure of YDII_ECOLI Northeast Structural Genomics Consortium Target ER29
To be Published
|
|
1S2C
 
 | | Crystal structures of prostaglandin D2 11-ketoreductase in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member C3, DIMETHYL SULFOXIDE, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
4HSO
 
 | | Crystal structure of S213G variant DAH7PS from Neisseria meningitidis | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Cross, P.J, Pietersma, A.L, Allison, T.M, Wilson-Coutts, S.M, Cochrane, F.C, Parker, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine.
Protein Sci., 22, 2013
|
|
2BT3
 
 | | AGAO in complex with Ruthenium-C4-wire at 1.73 angstroms | | Descriptor: | BIS[1H,1'H-2,2'-BIPYRIDINATO(2-)-KAPPA~2~N~1~,N~1'~]{3-[4-(1,10-DIHYDRO-1,10-PHENANTHROLIN-4-YL-KAPPA~2~N~1~,N~10~)BUTOXY]-N,N-DIMETHYLANILINATO(2-)}RUTHENIUM, COPPER (II) ION, GLYCEROL, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2005-05-26 | | Release date: | 2005-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Reversible Inhibition of Copper Amine Oxidase Activity by Channel-Blocking Ruthenium(II) and Rhenium(I) Molecular Wires.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BRC
 
 | | Structure of a Hsp90 Inhibitor bound to the N-terminus of Yeast Hsp90. | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
1RHK
 
 | |
1RSD
 
 | | DHNA complex with 3-(5-Amino-7-hydroxy-[1,2,3]triazolo[4,5-d]pyrimidin-2-yl)-N-[2-(2-hydroxymethyl-phenylsulfanyl)-benzyl]-benzamide | | Descriptor: | 3-(5-AMINO-7-HYDROXY-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-2-YL)-N-[2-(2-(HYDROXYMETHYL-PHENYLSULFANYL)-BENZYL]-BENZAMIDE, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2003-12-09 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Potent Inhibitors of Dihydroneopterin Aldolase Using CrystaLEAD High-Throughput X-ray Crystallographic Screening and Structure-Directed Lead Optimization.
J.Med.Chem., 47, 2004
|
|
1ZDR
 
 | | DHFR from Bacillus Stearothermophilus | | Descriptor: | GLYCEROL, SULFATE ION, dihydrofolate reductase | | Authors: | Kim, H.S, Damo, S.M, Lee, S.Y, Wemmer, D, Klinman, J.P. | | Deposit date: | 2005-04-14 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and hydride transfer mechanism of a moderate thermophilic dihydrofolate reductase from Bacillus stearothermophilus and comparison to its mesophilic and hyperthermophilic homologues.
Biochemistry, 44, 2005
|
|
1RHR
 
 | |
1ZP6
 
 | | Crystal Structure of Atu3015, a Putative Cytidylate Kinase from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR62 | | Descriptor: | SULFATE ION, hypothetical protein Atu3015 | | Authors: | Forouhar, F, Abashidze, M, Vorobiev, S.M, Kuzin, A, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-16 | | Release date: | 2005-05-24 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Atu3015, a Putative Cytidylate Kinase from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR62
To be Published
|
|
1R4H
 
 | | NMR Solution structure of the IIIc domain of GB Virus B IRES Element | | Descriptor: | 5'-R(*GP*GP*GP*CP*AP*AP*GP*CP*CP*C)-3' | | Authors: | Kaluarachchi, K, Thiviyanathan, V, Rijinbrand, R, Lemon, S.M, Gorenstein, D.G. | | Deposit date: | 2003-10-06 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites.
J.Mol.Biol., 343, 2004
|
|
1QBG
 
 | | CRYSTAL STRUCTURE OF HUMAN DT-DIAPHORASE (NAD(P)H OXIDOREDUCTASE) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Skelly, J.V, Sanderson, M.R, Suter, D.A, Baumann, U, Gregory, D.S, Bennett, M, Hobbs, S.M, Neidle, S. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human DT-diaphorase: a model for interaction with the cytotoxic prodrug 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954).
J.Med.Chem., 42, 1999
|
|
2BKU
 
 | | Kap95p:RanGTP complex | | Descriptor: | GTP-BINDING NUCLEAR PROTEIN RAN, GUANOSINE-5'-TRIPHOSPHATE, IMPORTIN BETA-1 SUBUNIT, ... | | Authors: | Lee, S.J, Matsuura, Y, Liu, S.M, Stewart, M. | | Deposit date: | 2005-02-21 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Nuclear Import Complex Dissociation by Rangtp
Nature, 435, 2005
|
|
1QSC
 
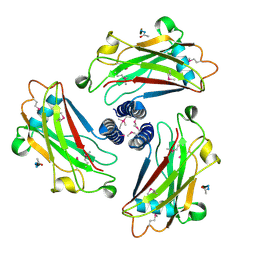 | | CRYSTAL STRUCTURE OF THE TRAF DOMAIN OF TRAF2 IN A COMPLEX WITH A PEPTIDE FROM THE CD40 RECEPTOR | | Descriptor: | CD40 RECEPTOR, TNF RECEPTOR ASSOCIATED FACTOR 2 | | Authors: | McWhirter, S.M, Pullen, S.S, Holton, J.M, Crute, J.J, Kehry, M.R, Alber, T. | | Deposit date: | 1999-06-20 | | Release date: | 1999-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic analysis of CD40 recognition and signaling by human TRAF2.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2BZX
 
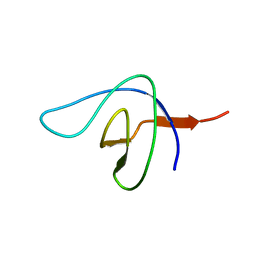 | |
2BRE
 
 | | STRUCTURE OF A HSP90 INHIBITOR BOUND TO THE N-TERMINUS OF YEAST HSP90. | | Descriptor: | 4-{4-[4-(3-AMINOPROPOXY)PHENYL]-1H-PYRAZOL-5-YL}-6-CHLOROBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
2C2V
 
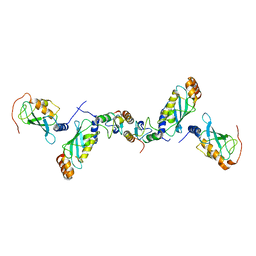 | | Crystal structure of the CHIP-UBC13-UEV1a complex | | Descriptor: | STIP1 homology and U box-containing protein 1, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Zhang, M, Roe, S.M, Pearl, L.H. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex.
Mol. Cell, 20, 2005
|
|
1S5R
 
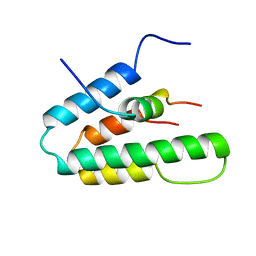 | | Solution Structure of HBP1 SID-mSin3A PAH2 Complex | | Descriptor: | Sin3a protein, high mobility group box transcription factor 1 | | Authors: | Swanson, K.A, Knoepfler, P.S, Huang, K, Kang, R.S, Cowley, S.M, Laherty, C.D, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2004-01-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1ZK7
 
 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase, ... | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-05-02 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
