6BBL
 
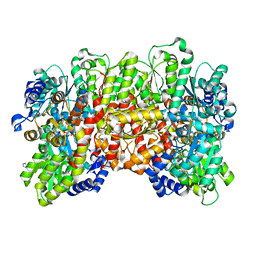 | | Crystal structure of the a-96Gln MoFe protein variant in the presence of the substrate acetylene | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Vertemara, J, Eilers, B.J, Karamatullah, D, Rasmussen, A.J, De Gioia, L, Zampella, G, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2017-10-18 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the nitrogenase molybdenum-iron protein with the substrate acetylene trapped near the active site.
J. Inorg. Biochem., 180, 2017
|
|
2PIG
 
 | | Crystal structure of ydcK from Salmonella cholerae at 2.38 A resolution. Northeast Structural Genomics target SCR6 | | Descriptor: | Putative transferase, ZINC ION | | Authors: | Benach, J, Chen, Y, Vorobiev, S.M, Seetharaman, J, Ho, C.K, Janjua, H, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of ydcK from Salmonella cholerae at 2.38 A resolution.
To be Published
|
|
2JKJ
 
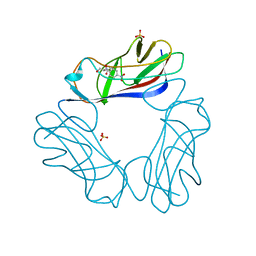 | | DraE Adhesin in complex with Chloramphenicol Succinate | | Descriptor: | CHLORAMPHENICOL, DR HEMAGGLUTININ STRUCTURAL SUBUNIT, SULFATE ION, ... | | Authors: | Pettigrew, D.M, Roversi, P, Davies, S.G, Russell, A.J, Lea, S.M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Structural Study of the Interaction between the Dr Haemagglutinin Drae and Derivatives of Chloramphenicol
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2KXG
 
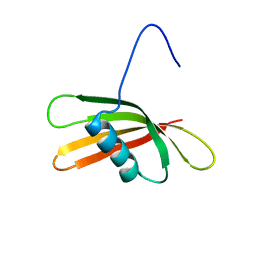 | | The solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) | | Descriptor: | Aspartic protease inhibitor | | Authors: | Headey, S.J, Macaskill, U.K, Wright, M, Claridge, J.K, Edwards, P.J.B, Farley, P.C, Christeller, J.T, Laing, W.A, Pascal, S.M. | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) and mutational analysis of pepsin inhibition.
J.Biol.Chem., 285, 2010
|
|
1CMX
 
 | |
6B25
 
 | |
2LKE
 
 | | Structures and Interaction Analyses of the Integrin Alpha-M Beta-2 Cytoplasmic Tails | | Descriptor: | Integrin alpha-M | | Authors: | Chua, G.L, Tang, X.Y, Amalraj, M, Bhattacharjya, S, Tan, S.M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures and interaction analyses of the integrin alphaMbeta2 cytoplasmic tails
J.Biol.Chem., 2011
|
|
8FCK
 
 | | Structure of the vertebrate augmin complex | | Descriptor: | HAUS augmin like complex subunit 2 L homeolog, Green fluorescent protein chimera, HAUS augmin like complex subunit 4 L homeolog, ... | | Authors: | Travis, S.M, Huang, W, Zhang, R, Petry, S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.88 Å) | | Cite: | Integrated model of the vertebrate augmin complex.
Nat Commun, 14, 2023
|
|
3ZUI
 
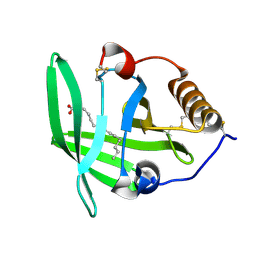 | | OMCI in complex with palmitoleic acid | | Descriptor: | COMPLEMENT INHIBITOR, PALMITOLEIC ACID | | Authors: | Roversi, P, Maillet, I, Togbe, D, Couillin, I, Quesniaux, V.F.J, Teixeira, M, Ahmat, N, Lissina, O, Boland, W, Ploss, K, Caesar, J.J.E, Leonhartsberger, S, Ryffel, B, Lea, S.M, Nunn, M.A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Bifunctional Lipocalin Ameliorates Murine Immune Complex-Induced Acute Lung Injury.
J.Biol.Chem., 288, 2013
|
|
3ZUO
 
 | | OMCI in complex with leukotriene B4 | | Descriptor: | COMPLEMENT INHIBITOR, LEUKOTRIENE B4 | | Authors: | Roversi, P, Maillet, I, Togbe, D, Couillin, I, Quesniaux, V.F.J, Teixeira, M, Ahmat, N, Lissina, O, Boland, W, Ploss, K, Caesar, J.J.E, Leonhartsberger, S, Ryffel, B, Lea, S.M, Nunn, M.A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Bifunctional Lipocalin Ameliorates Murine Immune Complex-Induced Acute Lung Injury.
J.Biol.Chem., 288, 2013
|
|
6BYS
 
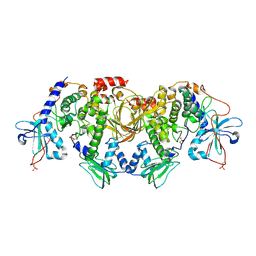 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PRKAc alpha | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.75 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
6BSP
 
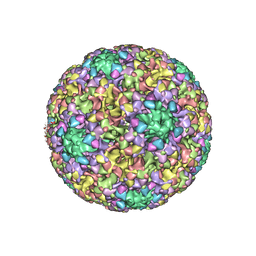 | | High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitope on Human Papillomavirus 16 | | Descriptor: | Major capsid protein L1, U4 Heavy chain, U4 Light chain | | Authors: | Guan, J, Bywaters, S.M, Brendle, S.A, Ashley, R.E, Makhov, A.M, Conway, J.F, Christenson, N.D, Hafenstein, S. | | Deposit date: | 2017-12-04 | | Release date: | 2018-02-14 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | High-Resolution Structure Analysis of Antibody V5 and U4 Conformational Epitopes on Human Papillomavirus 16.
Viruses, 9, 2017
|
|
2LF7
 
 | |
1BC2
 
 | | ZN-DEPENDENT METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS | | Descriptor: | METALLO-BETA-LACTAMASE II, SULFATE ION, ZINC ION | | Authors: | Fabiane, S.M, Sutton, B.J. | | Deposit date: | 1997-04-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the zinc-dependent beta-lactamase from Bacillus cereus at 1.9 A resolution: binuclear active site with features of a mononuclear enzyme.
Biochemistry, 37, 1998
|
|
2UWN
 
 | | Crystal structure of Human Complement Factor H, SCR domains 6-8 (H402 risk variant), in complex with ligand. | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Prosser, B.E, Johnson, S, Roversi, P, Herbert, A.P, Blaum, B.S, Tyrrell, J, Jowitt, T.A, Clark, S.J, Terelli, E, Uhrin, D, Barlow, P.N, Sim, R.B, Day, A.J, Lea, S.M. | | Deposit date: | 2007-03-22 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Complement Factor H Linked Age-Related Macular Degeneration.
J.Exp.Med., 204, 2007
|
|
2MZC
 
 | | Metal Binding of Glutaredoxins | | Descriptor: | Glutaredoxin, SILVER ION | | Authors: | Bilinovich, S.M, Caporoso, J.A, Taraboletti, A, Duangjumpa, N, Panzner, M.J, Prokop, J.W, Shriver, L.P, Leeper, T.C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal Binding of Glutaredoxins
To be Published
|
|
2O4W
 
 | | T4 lysozyme circular permutant | | Descriptor: | CHLORIDE ION, Lysozyme | | Authors: | Llinas, M, Crowder, S.M, Echols, N, Alber, T, Marqusee, S. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring subdomain cooperativity in T4 lysozyme I: Structural and energetic studies of a circular permutant and protein fragment.
Protein Sci., 16, 2007
|
|
1DQD
 
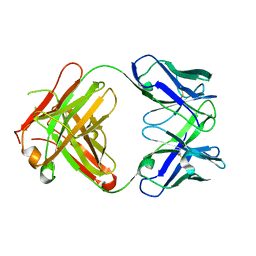 | | CRYSTAL STRUCTURE OF FAB HGR-2 F6, A COMPETITIVE ANTAGONIST OF THE GLUCAGON RECEPTOR | | Descriptor: | FAB HGR-2 F6 | | Authors: | Wright, L.M, Brzozowski, A.M, Hubbard, R.E, Pike, A.C.W, Roberts, S.M, Skovgaard, R.N, Svendsen, I, Vissing, H, Bywater, R.P. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Fab hGR-2 F6, a competitive antagonist of the glucagon receptor.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2UXV
 
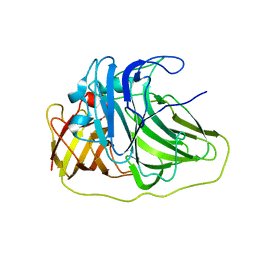 | | SufI Protein from Escherichia Coli | | Descriptor: | PROTEIN SUFI | | Authors: | Tarry, M.J, Roversi, P, Sargent, F, Berks, B.C, Lea, S.M. | | Deposit date: | 2007-03-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The Escherichia Coli Cell Division Protein and Model Tat Substrate Sufi (Ftsp) Localizes to the Septal Ring and Has a Multicopper Oxidase-Like Structure.
J.Mol.Biol., 386, 2009
|
|
4CGL
 
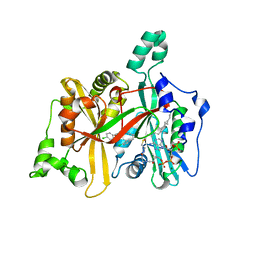 | | Leishmania major N-myristoyltransferase in complex with an aminoacylpyrrolidine inhibitor | | Descriptor: | (3R)-3-azanyl-4-(4-chlorophenyl)-1-[(3S,4R)-3-(4-chlorophenyl)-4-(hydroxymethyl)pyrrolidin-1-yl]butan-1-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
2K73
 
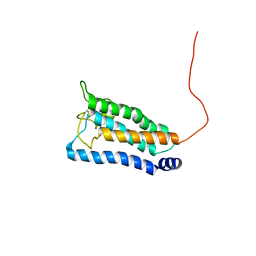 | | Solution NMR structure of integral membrane protein DsbB | | Descriptor: | Disulfide bond formation protein B | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
2K6T
 
 | | Solution structure of the relaxin-like factor | | Descriptor: | Insulin-like 3 A chain, Insulin-like 3 B chain | | Authors: | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | Deposit date: | 2008-07-23 | | Release date: | 2008-12-16 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
2KKF
 
 | | Solution structure of MLL CXXC domain in complex with palindromic CPG DNA | | Descriptor: | 5'-D(*CP*CP*CP*TP*GP*CP*GP*CP*AP*GP*GP*G)-3', Histone-lysine N-methyltransferase HRX, ZINC ION | | Authors: | Cierpicki, T, Riesbeck, J.E, Grembecka, J.E, Lukasik, S.M, Popovic, R, Omonkowska, M, Shultis, D.S, Zeleznik-Le, N.J, Bushweller, J.H. | | Deposit date: | 2009-06-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2K74
 
 | | Solution NMR structure of DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, UBIQUINONE-2 | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
6A4L
 
 | | AcrR from Mycobacterium tuberculosis | | Descriptor: | SODIUM ION, SULFATE ION, TetR family transcriptional regulator | | Authors: | Kang, S.M. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2020-04-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of AcrR from Mycobacterium tuberculosis reveals a one-component transcriptional regulation mechanism.
Febs Open Bio, 9, 2019
|
|
