6NTN
 
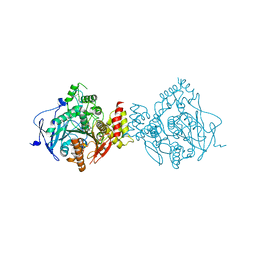 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-230 in Complex with the Reactivator, HI-6 | | Descriptor: | (S)-N-[(1E)-1-(diethylamino)ethylidene]-P-methylphosphonamidic fluoride, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
6NND
 
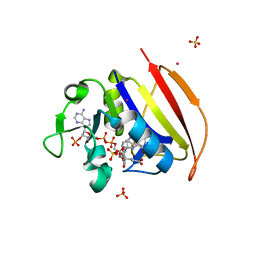 | | Structure of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and dihydrofolate | | Descriptor: | COBALT (II) ION, DIHYDROFOLIC ACID, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6NTO
 
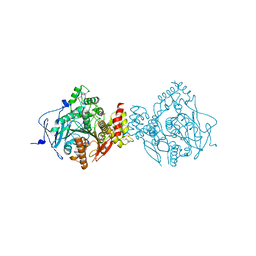 | | Crystal Structure of Recombinant Human Acetylcholinesterase Inhibited by A-230 | | Descriptor: | (S)-N-[(1E)-1-(diethylamino)ethylidene]-P-methylphosphonamidic fluoride, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, Guelta, M.A, Height, J.J, Pegan, S.D. | | Deposit date: | 2019-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Insights into inhibition of human acetylcholinesterase by Novichok, A-series Nerve Agents
To Be Published
|
|
1NGJ
 
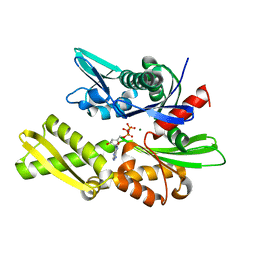 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
6F0O
 
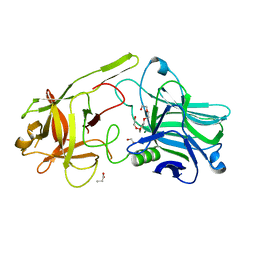 | | Botulinum neurotoxin A3 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, Bontoxilysin A, PENTAETHYLENE GLYCOL, ... | | Authors: | Davies, J.R, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
1KJQ
 
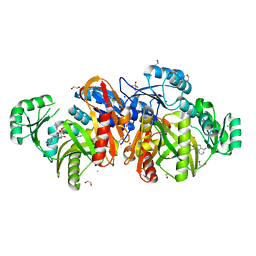 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-ADP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-05 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
8G2D
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with tylosin, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8G2C
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with tylosin, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.65A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
6S1M
 
 | | Human polymerase delta holoenzyme Conformer 1 | | Descriptor: | DNA polymerase delta catalytic subunit, DNA polymerase delta subunit 2, DNA polymerase delta subunit 3, ... | | Authors: | Lancey, C, Hamdan, S.M, De Biasio, A. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structure of the processive human Pol delta holoenzyme.
Nat Commun, 11, 2020
|
|
6P9E
 
 | | Crystal structure of IL-36gamma complexed to A-552 | | Descriptor: | (2S)-2-{[4-(3-amino-4-methylphenyl)-6-methylpyrimidin-2-yl]oxy}-3-methoxy-3,3-diphenylpropanoic acid, Interleukin-36 gamma | | Authors: | Argiriadi, M.A, Todorovic, T, Su, Z, Putman, B, Kakavas, S.J, Salte, K.M, McDonald, H.A, Wetter, J.B, Paulsboe, S.E, Sun, Q, Gerstein, C.E, Medina, L, Sielaff, B, Sadhukhan, R, Stockmann, H, Richardson, P.L, Qiu, W, Henry, R.F, Herold, J.M, Shotwell, J.B, McGaraughty, S.P, Honore, P, Gopalakrishnan, S.M, Sun, C.C, Scott, V.E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecule IL-36 gamma Antagonist as a Novel Therapeutic Approach for Plaque Psoriasis.
Sci Rep, 9, 2019
|
|
1KS5
 
 | | Structure of Aspergillus niger endoglucanase | | Descriptor: | Endoglucanase A | | Authors: | Khademi, S, Zhang, D, Swanson, S.M, Wartenberg, A, Witte, C, Meyer, E.F. | | Deposit date: | 2002-01-10 | | Release date: | 2003-01-21 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6RQJ
 
 | | Structure of human complement C5 complexed with tick inhibitors OmCI, RaCI1 and CirpT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5, Complement inhibitor, ... | | Authors: | Reichhardt, M.P, Johnson, S, Lea, S.M. | | Deposit date: | 2019-05-15 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An inhibitor of complement C5 provides structural insights into activation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6DZQ
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Dick, B.L, Morrison, C.N, Cohen, S.M. | | Deposit date: | 2018-07-05 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Activity Relationships in Metal-Binding Pharmacophores for Influenza Endonuclease.
J. Med. Chem., 61, 2018
|
|
6E3M
 
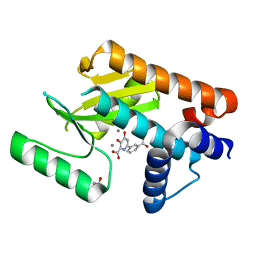 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 6-(3-carboxyphenyl)-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-carboxyphenyl)-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Morrison, C.N, Dick, B.L, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
6E3N
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-4-oxo-6-(o-tolyl)-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 3-hydroxy-6-(2-methylphenyl)-4-oxo-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Morrison, C.N, Dick, B.L, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
6E3O
 
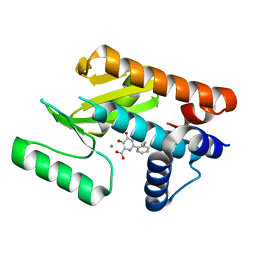 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 6-(2-ethylphenyl)-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 6-(2-ethylphenyl)-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Morrison, C.N, Dick, B.L, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
6E3P
 
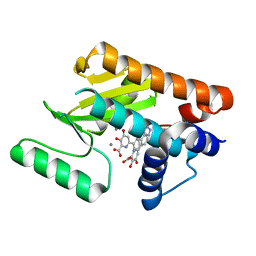 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 6-(3-(1H-tetrazol-5-yl)phenyl)-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 3-hydroxy-4-oxo-6-[3-(1H-tetrazol-5-yl)phenyl]-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Morrison, C.N, Dick, B.L, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
6SA4
 
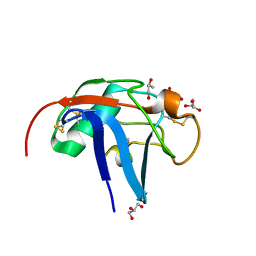 | | SALSA / DMBT1 / GP340 SRCR domain 1 | | Descriptor: | CHLORIDE ION, Deleted in malignant brain tumors 1 protein, GLYCEROL, ... | | Authors: | Reichhardt, M.P, Lea, S.M, Johnson, S. | | Deposit date: | 2019-07-16 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.
Life Sci Alliance, 3, 2020
|
|
1NGD
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
6E4C
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-6-(2-methyl-4-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)phenyl)-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-[2-methyl-4-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)phenyl]-4-oxo-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Morrison, C.N, Dick, B.L, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-17 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
1NF3
 
 | | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6 | | Descriptor: | G25K GTP-binding protein, placental isoform, MAGNESIUM ION, ... | | Authors: | Garrard, S.M, Capaldo, C.T, Gao, L, Rosen, M.K, Macara, I.G, Tomchick, D.R. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6
Embo J., 22, 2003
|
|
1LU5
 
 | | 2.4 Angstrom Crystal Structure of the Asymmetric Platinum Complex {Pt(ammine)(cyclohexylamine)}2+ Bound to a Dodecamer DNA Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', CIS-(AMMINE)(CYCLOHEXYLAMINE)PLATINUM(II) COMPLEX | | Authors: | Silverman, A.P, Bu, W, Cohen, S.M, Lippard, S.J. | | Deposit date: | 2002-05-21 | | Release date: | 2002-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4-A Crystal Structure of the Asymmetric Platinum Complex {Pt(ammine)(cyclohexylamine)}2+ Bound to
a Dodecamer DNA Duplex
J.Biol.Chem., 277, 2002
|
|
1LCY
 
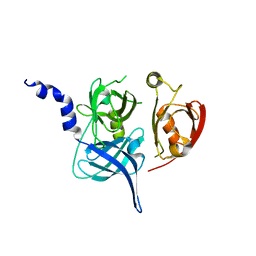 | | Crystal Structure of the Mitochondrial Serine Protease HtrA2 | | Descriptor: | HtrA2 serine protease | | Authors: | Li, W, Srinivasula, S.M, Chai, J, Li, P, Wu, J.W, Zhang, Z, Alnemri, E.S, Shi, Y. | | Deposit date: | 2002-04-07 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi.
Nat.Struct.Biol., 9, 2002
|
|
2J9T
 
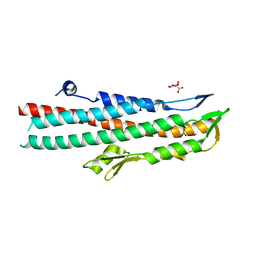 | | BipD of Burkholderia Pseudomallei | | Descriptor: | BORIC ACID, CITRATE ANION, MEMBRANE ANTIGEN | | Authors: | Johnson, S, Roversi, P, Field, T, Deane, J.E, Galyov, E, Lea, S.M. | | Deposit date: | 2006-11-16 | | Release date: | 2006-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Self-Chaperoning of the Type III Secretion System Needle Tip Proteins Ipad and Bipd.
J.Biol.Chem., 282, 2007
|
|
6SD5
 
 | | Structure of the RBM2 inner ring of Salmonella flagella MS-ring protein FliF with 22-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
