2QDQ
 
 | | Crystal structure of the talin dimerisation domain | | Descriptor: | Talin-1 | | Authors: | Gingras, A.R, Putz, N.S.M, Bate, N, Barsukov, I.L, Critchley, D.R.C. | | Deposit date: | 2007-06-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the C-terminal actin-binding domain of talin.
Embo J., 27, 2008
|
|
5BPA
 
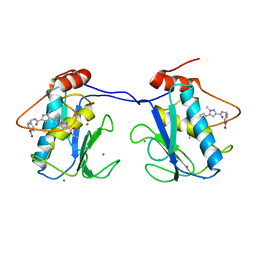 | |
4E1L
 
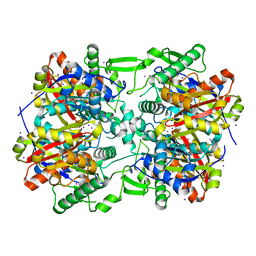 | | Crystal structure of Acetoacetyl-CoA thiolase (thlA2) from Clostridium difficile | | Descriptor: | Acetoacetyl-CoA thiolase 2, IODIDE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: |
|
|
2YKD
 
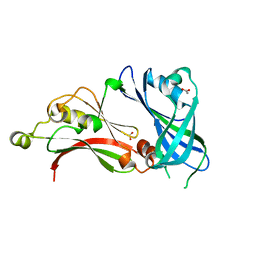 | | Structure of the matrix protein from human respiratory syncytial virus | | Descriptor: | ACETATE ION, MATRIX PROTEIN | | Authors: | McPhee, H.K, Carlisle, J.L, Beeby, A, Money, V.A, Watson, S.M.D, Yeo, R.P, Sanderson, J.M. | | Deposit date: | 2011-05-26 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Influence of Lipids on the Interfacial Disposition of Respiratory Syncytical Virus Matrix Protein.
Langmuir, 27, 2011
|
|
4DBL
 
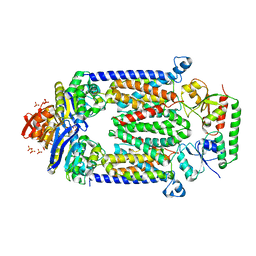 | | Crystal structure of E159Q mutant of BtuCDF | | Descriptor: | PHOSPHATE ION, SULFATE ION, Vitamin B12 import ATP-binding protein BtuD, ... | | Authors: | Korkhov, V.M, Mireku, S.M, Hvorup, R.N, Locher, K.P. | | Deposit date: | 2012-01-16 | | Release date: | 2012-03-07 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | Asymmetric states of vitamin B12 transporter BtuCD are not discriminated by its cognate substrate binding protein BtuF.
Febs Lett., 586, 2012
|
|
2X35
 
 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | PEREGRIN | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4G8V
 
 | | Crystal structure of Ribonuclease A in complex with 5a | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
4X25
 
 | |
2X4W
 
 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | FORMIC ACID, HISTONE H3.2, PEREGRIN | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2X4Y
 
 | | Molecular basis of Histone H3K36me3 recognition by the PWWP domain of BRPF1. | | Descriptor: | HISTONE H3.2, PEREGRIN, SULFATE ION | | Authors: | Vezzoli, A, Bonadies, N, Allen, M.D, Freund, S.M.V, Santiveri, C.M, Kvinlaug, B, Huntly, B.J.P, Gottgens, B, Bycroft, M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-21 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis of Histone H3K36Me3 Recognition by the Pwwp Domain of Brpf1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4X1L
 
 | |
3Q58
 
 | | Structure of N-acetylmannosamine-6-Phosphate Epimerase from Salmonella enterica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-27 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: |
|
|
4WZP
 
 | | Ser65 phosphorylated ubiquitin, major conformation | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Wauer, T, Wagstaff, J, Freund, S.M.V, Komander, D. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis.
Embo J., 34, 2015
|
|
4X1M
 
 | |
4FMP
 
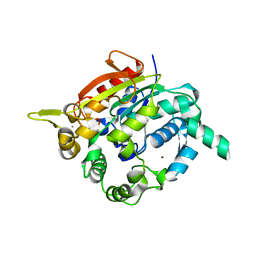 | | Crystal structure of thermostable, organic-solvent tolerant lipase from Geobacillus sp. strain ARM | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Nisbar, N.D, Rahman, R.N.Z.R.A, Ali, M.S.M, Leow, A.T.C. | | Deposit date: | 2012-06-18 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of novel ARM lipase and elucidation of its space-grown crystal structure
Thesis, 2013
|
|
2Y9T
 
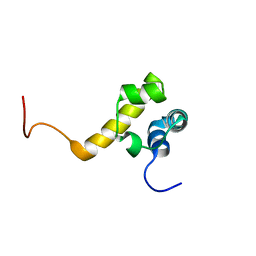 | |
4ZP7
 
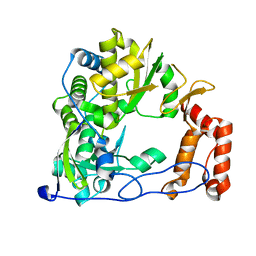 | | Coxsackievirus B3 Polymerase - F364V mutant | | Descriptor: | Genome polyprotein | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
2PTX
 
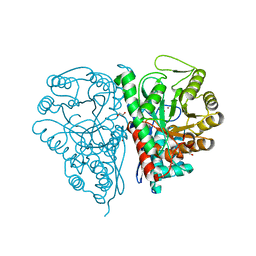 | | Crystal Structure of the T. brucei enolase complexed with sulphate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, SULFATE ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
4ZPC
 
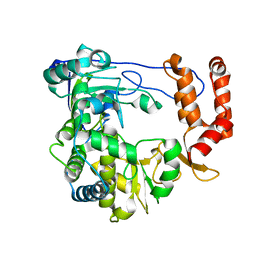 | | Coxsackievirus B3 Polymerase - A341G mutant | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Peersen, O.B, McDonald, S.M. | | Deposit date: | 2015-05-07 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo.
J.Biol.Chem., 291, 2016
|
|
4FMX
 
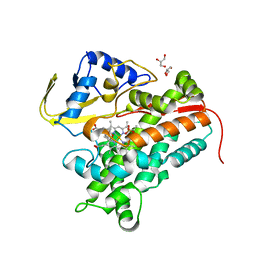 | | Crystal Structure of Substrate-Bound P450cin | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, GLYCEROL, P450cin, ... | | Authors: | Madrona, Y, Tripathi, S.M, Huiying, L, Poulos, T.L. | | Deposit date: | 2012-06-18 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
4FA3
 
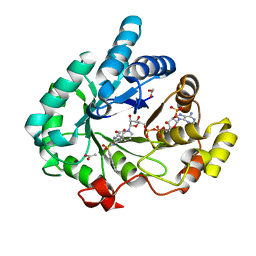 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with (R)-1-(naphthalen-2-ylsulfonyl)piperidine-3-carboxylic acid (86) | | Descriptor: | (3R)-1-(naphthalen-2-ylsulfonyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
4XXO
 
 | |
2Y9U
 
 | | Structural basis of p63a SAM domain mutants involved in AEC syndrome | | Descriptor: | SULFATE ION, TUMOR PROTEIN 63 | | Authors: | Sathyamurthy, A, Freund, S.M.V, Johnson, C.M, Allen, M.D. | | Deposit date: | 2011-02-16 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of P63Alpha Sam Domain Mutants Involved in Aec Syndrome.
FEBS J., 278, 2011
|
|
3PXM
 
 | | Reduced sweetness of a monellin (MNEI) mutant results from increased protein flexibility and disruption of a distant poly-(L-proline) II helix | | Descriptor: | Monellin chain B/Monellin chain A chimeric protein | | Authors: | Hobbs, J.R, Templeton, C.M, Pour, S.O, Blanch, E.W, Munger, S.M, Conn, G.L. | | Deposit date: | 2010-12-10 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reduced Sweetness of a Monellin (MNEI) Mutant Results from Increased Protein Flexibility and Disruption of a Distant Poly-(L-Proline) II Helix.
Chem Senses, 36, 2011
|
|
4YI3
 
 | | Crystal structure of Gpb in complex with 4a | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Stravodimos, G.A, Leonidas, D.D. | | Deposit date: | 2015-02-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycogen phosphorylase as a target for type 2 diabetes: synthetic, biochemical, structural and computational evaluation of novel N-acyl-N -( beta-D-glucopyranosyl) urea inhibitors.
Curr Top Med Chem, 15, 2015
|
|
