7AGI
 
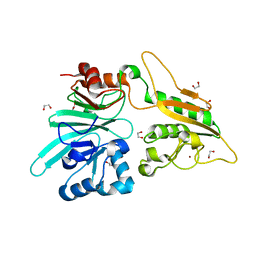 | | The structure of Artemis variant H35D | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7B2X
 
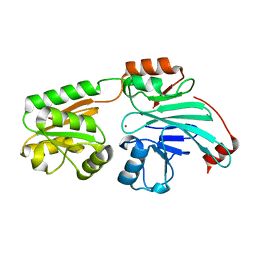 | | Crystal structure of human 5' exonuclease Appollo H61Y variant | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-28 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
7B9B
 
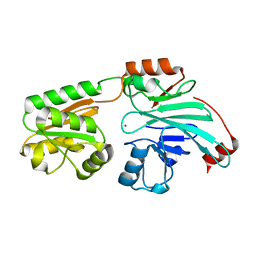 | | Crystal structure of human 5' exonuclease Appollo APO form | | Descriptor: | 5' exonuclease Apollo, NICKEL (II) ION | | Authors: | Newman, J.A, Baddock, H.T, Mukhopadhyay, S.M.M, Burgess-Brown, N.A, von Delft, F, Arrowshmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-12-14 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A phosphate binding pocket is a key determinant of exo- versus endo-nucleolytic activity in the SNM1 nuclease family.
Nucleic Acids Res., 49, 2021
|
|
7BUK
 
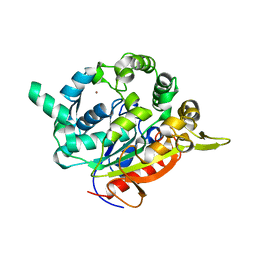 | | T1 lipase mutant - 5M (D43E/T118N/E226D/E250L/N304E) | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Ishak, S.N.H, Rahman, R.N.Z.R.A, Ali, M.S.M, Leow, A.T.C, Kamarudin, N.H.A. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Structure elucidation and docking analysis of 5M mutant of T1 lipase Geobacillus zalihae.
Plos One, 16, 2021
|
|
5TED
 
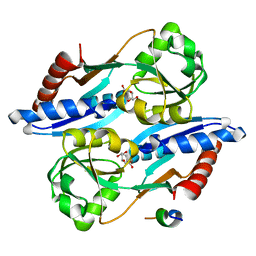 | | Effector binding domain of QuiR in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, His Tag peptide, Lmo0488 protein | | Authors: | Prezioso, S.M, Christendat, D. | | Deposit date: | 2016-09-21 | | Release date: | 2017-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Shikimate Induced Transcriptional Activation of Protocatechuate Biosynthesis Genes by QuiR, a LysR-Type Transcriptional Regulator, in Listeria monocytogenes.
J. Mol. Biol., 430, 2018
|
|
5TJK
 
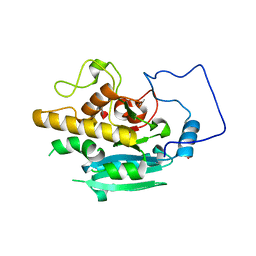 | | Crystal structure of GTA + A trisaccharide (native) | | Descriptor: | GLYCEROL, Histo-blood group ABO system transferase, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | Authors: | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | Deposit date: | 2016-10-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
5TJN
 
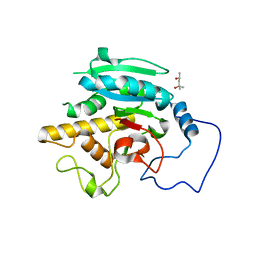 | | Crystal structure of GTB + B trisaccharide (native) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histo-blood group ABO system transferase, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | Authors: | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | Deposit date: | 2016-10-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
5TJL
 
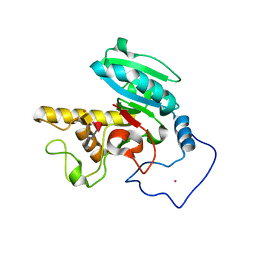 | | Crystal structure of GTA + A trisaccharide (mercury derivative) | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | Authors: | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | Deposit date: | 2016-10-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
5TJO
 
 | | Crystal structure of GTB + B trisaccharide (mercury derivative) | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]octyl beta-D-galactopyranoside | | Authors: | Legg, M.S.G, Gagnon, S.M.L, Evans, S.V. | | Deposit date: | 2016-10-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | High-resolution crystal structures and STD NMR mapping of human ABO(H) blood group glycosyltransferases in complex with trisaccharide reaction products suggest a molecular basis for product release.
Glycobiology, 27, 2017
|
|
5TXL
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG) P*CP*GP*CP*CP*GP)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5U0K
 
 | | C-terminal ankyrin repeats from human liver-type glutaminase (GAB/LGA) | | Descriptor: | Glutaminase liver isoform, mitochondrial | | Authors: | Ferreira, I.M, Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5TXP
 
 | | STRUCTURE OF Q151M complex (A62V, V75I, F77L, F116Y, Q151M) mutant HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DDATP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5UF6
 
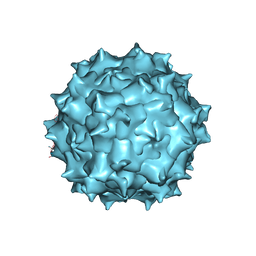 | | The 2.8 A Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, capsid protein VP1 | | Authors: | Xie, Q, Spear, J.M, Noble, A.J, Sousa, D.R, Meyer, N.L, Davulcu, O, Zhang, F, Linhardt, R.J, Stagg, S.M, Chapman, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The 2.8 angstrom Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparinoid Pentasaccharide.
Mol Ther Methods Clin Dev, 5, 2017
|
|
5TXN
 
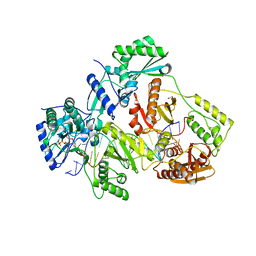 | | STRUCTURE OF Q151M MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
5TZL
 
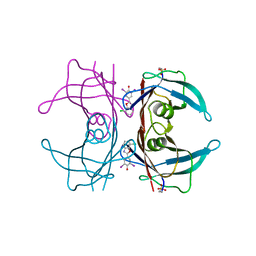 | | Structure of transthyretin in complex with the kinetic stabilizer 201 | | Descriptor: | 4-(7-chloro-1,3-benzoxazol-2-yl)-2,6-diiodophenol, Transthyretin | | Authors: | Connelly, S, Mortenson, D.E, Choi, S, Wilson, I.A, Powers, E.T, Kelly, J.W, Johnson, S.M. | | Deposit date: | 2016-11-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Semi-quantitative models for identifying potent and selective transthyretin amyloidogenesis inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TZY
 
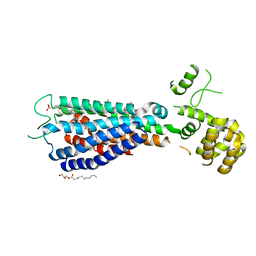 | | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R)-3-cyclopropyl-3-[(2R)-2-(1-{(1S)-1-[5-fluoro-2-(trifluoromethoxy)phenyl]ethyl}piperidin-4-yl)-3,4-dihydro-2H-1-benzopyran-7-yl]-2-methylpropanoic acid, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5TSQ
 
 | | Crystal structure of IUnH from Leishmania braziliensis in complex with D-Ribose | | Descriptor: | CALCIUM ION, IUnH, beta-D-ribofuranose | | Authors: | Bachega, J.F.R, Timmers, L.F.S.M, Dalberto, P.F, Martinelli, L, Pinto, A.F.M, Basso, L.A, Norberto de Souza, O, Santos, D.S. | | Deposit date: | 2016-10-31 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of IUnH from Leishmania braziliensis in complex with D-Ribose
To Be Published
|
|
5TY3
 
 | | Crystal structure of K72A variant of Human Cytochrome c | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Mou, T.C, Nold, S.M, Lei, H, Sprang, S.R, Bowler, B.E. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effect of a K72A Mutation on the Structure, Stability, Dynamics, and Peroxidase Activity of Human Cytochrome c.
Biochemistry, 56, 2017
|
|
