6ZIS
 
 | | Crystal structure of a CGRP receptor ectodomain heterodimer with bound high affinity inhibitor | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, N-{(1S)-5-amino-1-[(4-pyridin-4-ylpiperazin-1-yl)carbonyl]pentyl}-3,5-dibromo-Nalpha-{[4-(2-oxo-1,4-dihydroquinazolin-3 (2H)-yl)piperidin-1-yl]carbonyl}-D-tyrosinamide, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Drug Discovery ofN-((R)-3-(7-Methyl-1H-indazol-5-yl)-1-oxo-1-(((S)-1-oxo-3-(piperidin-4-yl)-1-(4-(pyridin-4-yl)piperazin-1-yl)propan-2-yl)amino)propan-2-yl)-2'-oxo-1',2'-dihydrospiro[piperidine-4,4'-pyrido[2,3-d][1,3]oxazine]-1-carboxamide (HTL22562): A Calcitonin Gene-Related Peptide Receptor Antagonist for Acute Treatment of Migraine.
J.Med.Chem., 63, 2020
|
|
4CHH
 
 | | N-terminal domain of yeast PIH1p | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN INTERACTING WITH HSP90 1 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CGU
 
 | | Full length Tah1 bound to yeast PIH1 and HSP90 peptide SRMEEVD | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA, PROTEIN INTERACTING WITH HSP90 1, ... | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-05-14 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
1LO1
 
 | | ESTROGEN RELATED RECEPTOR 2 DNA BINDING DOMAIN IN COMPLEX WITH DNA | | Descriptor: | 5'-D(*CP*GP*TP*GP*AP*CP*CP*TP*TP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*AP*AP*GP*GP*TP*CP*AP*CP*G)-3', Steroid hormone receptor ERR2, ... | | Authors: | Gearhart, M.D, Holmbeck, S.M.A, Evans, R.M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-05-05 | | Release date: | 2003-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Monomeric Complex of Human Orphan Estrogen Related Receptor-2 with DNA: A Pseudo-dimer Interface Mediates Extended Half-site Recognition
J.Mol.Biol., 327, 2003
|
|
1ZHV
 
 | | X-ray Crystal Structure Protein Atu0741 from Agobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR8. | | Descriptor: | hypothetical protein Atu0741 | | Authors: | Vorobiev, S.M, Kuzin, A, Abashidze, M, Forouhar, F, Xio, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-26 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of hypothetical protein Atu071 from Agobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR8.
To be Published
|
|
4CUF
 
 | | Human Notch1 EGF domains 11-13 mutant T466S | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1RXR
 
 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE RETINOID X RECEPTOR DNA BINDING DOMAIN, NMR, 20 STRUCTURE | | Descriptor: | RETINOIC ACID RECEPTOR-ALPHA, ZINC ION | | Authors: | Holmbeck, S.M.A, Foster, M.P, Casimiro, D.R, Sem, D.S, Dyson, H.J, Wright, P.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the retinoid X receptor DNA-binding domain.
J.Mol.Biol., 281, 1998
|
|
4BF3
 
 | | ErpC, a member of the complement regulator acquiring family of surface proteins from Borrelia burgdorfei, possesses an architecture previously unseen in this protein family. | | Descriptor: | 1,2-ETHANEDIOL, ERPC | | Authors: | Caesar, J.J.E, Johnson, S, Kraiczy, P, Lea, S.M. | | Deposit date: | 2013-03-14 | | Release date: | 2013-06-05 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Erpc, a Member of the Complement Regulator-Acquiring Family of Surface Proteins from Borrelia Burgdorferi, Possesses an Architecture Previously Unseen in This Protein Family.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2BW2
 
 | | BofC from Bacillus subtilis | | Descriptor: | BYPASS OF FORESPORE C | | Authors: | Patterson, H.M, Brannigan, J.A, Cutting, S.M, Wilson, K.S, Wilkinson, A.J, Ab, E, Diercks, T, Folkers, G.E, de Jong, R.N, Truffault, V, Kaptein, R. | | Deposit date: | 2005-07-08 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of bypass of forespore C, an intercompartmental signaling factor during sporulation in Bacillus.
J. Biol. Chem., 280, 2005
|
|
2BY7
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BYA
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY6
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY9
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY8
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BY5
 
 | | Is radiation damage dependent on the dose-rate used during macromolecular crystallography data collection | | Descriptor: | BENZAMIDINE, CALCIUM ION, CATIONIC TRYPSIN, ... | | Authors: | Leiros, H.-K.S, Timmins, J, Ravelli, R.B.G, McSweeney, S.M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-02-06 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Is Radiation Damage Dependent on the Dose-Rate Used During Macromolecular Crystallography Data Collection?
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4BFG
 
 | | Structure of the extracellular portion of mouse CD200R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, AZIDE ION, ... | | Authors: | Hatherley, D, Lea, S.M, Johnson, S, Barclay, A.N. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of Cd200/Cd200 Receptor Family and Implications for Topology, Regulation, and Evolution
Structure, 21, 2013
|
|
1ZP6
 
 | | Crystal Structure of Atu3015, a Putative Cytidylate Kinase from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR62 | | Descriptor: | SULFATE ION, hypothetical protein Atu3015 | | Authors: | Forouhar, F, Abashidze, M, Vorobiev, S.M, Kuzin, A, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-16 | | Release date: | 2005-05-24 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Atu3015, a Putative Cytidylate Kinase from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR62
To be Published
|
|
6YYE
 
 | | TREM2 extracellular domain (19-131) in complex with single-chain variable fragment (scFv-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TREM2 Single chain variable 2, Triggering receptor expressed on myeloid cells 2 | | Authors: | Szykowska, A, Preger, C, Krojer, T, Mukhopadhyay, S.M.M, McKinley, G, Graslund, S, Wigren, E, Persson, H, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Di Daniel, E, Burgess-Brown, N, Bullock, A. | | Deposit date: | 2020-05-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
4B6F
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (4-phenoxyphenyl)methylazanium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, ... | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
1ZBA
 
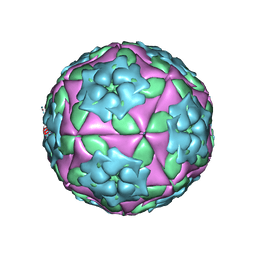 | | Foot-and-Mouth Disease virus serotype A1061 complexed with oligosaccharide receptor. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Coat protein VP1, Coat protein VP2, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
4AVB
 
 | | Crystal structure of protein lysine acetyltransferase Rv0998 in complex with acetyl CoA and cAMP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Lee, H.J, Lang, P.T, Fortune, S.M, Sassetti, C.M, Alber, T. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclic AMP Regulation of Protein Lysine Acetylation in Mycobacterium Tuberculosis.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4B73
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-amino-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
1Z94
 
 | | X-Ray Crystal Structure of Protein CV1439 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR12. | | Descriptor: | conserved hypothetical protein | | Authors: | Kuzin, A, Vorobiev, S.M, Yong, W, Forouhar, F, Xio, R, Ma, L.-C, Acton, T, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hypothetical Protein CV1439 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CVR12.
To be Published
|
|
1ZBE
 
 | | Foot-and Mouth Disease Virus Serotype A1061 | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Newman, J.W, Curry, S, Najjam, S, Jackson, T, Blakemore, W, Lea, S.M, Miller, L, Burman, A, King, A.M, Stuart, D.I. | | Deposit date: | 2005-04-08 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Foot-and-mouth disease virus serotype A1061 alone and complexed with oligosaccharide receptor: receptor conservation in the face of antigenic variation.
J.Gen.Virol., 86, 2005
|
|
1ZC6
 
 | | Crystal Structure of Putative N-acetylglucosamine Kinase from Chromobacterium violaceum. Northeast Structural Genomics Target Cvr23. | | Descriptor: | probable N-acetylglucosamine kinase | | Authors: | Vorobiev, S.M, Kuzin, A, Forouhar, F, Abashidze, M, Acton, T.B, Xiao, R, Ma, L.-C, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-11 | | Release date: | 2005-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Putative N-acetylglucosamine Kinase from Chromobacterium violaceum
To be Published
|
|
