3KZB
 
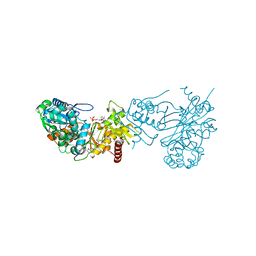 | |
3PQB
 
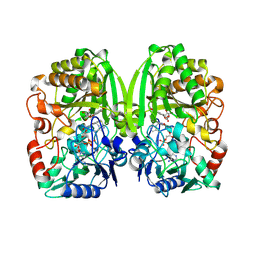 | | The crystal structure of pregilvocarcin in complex with GilR, an oxidoreductase that catalyzes the terminal step of gilvocarcin biosynthesis | | Descriptor: | (1R)-1,4-anhydro-6-deoxy-1-[(6R)-8-ethenyl-1,6-dihydroxy-10,12-dimethoxy-6H-dibenzo[c,h]chromen-4-yl]-D-galactitol, FLAVIN-ADENINE DINUCLEOTIDE, Putative oxidoreductase | | Authors: | Noinaj, N, Bosserman, M.A, Schickli, M.A, Kharel, M.K, Rohr, J, Buchanan, S.K. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | The Crystal Structure and Mechanism of an Unusual Oxidoreductase, GilR, Involved in Gilvocarcin V Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
8TVL
 
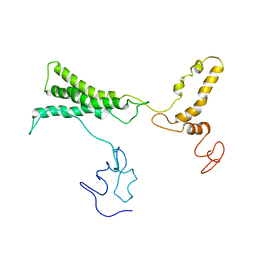 | |
3Q83
 
 | |
8TWE
 
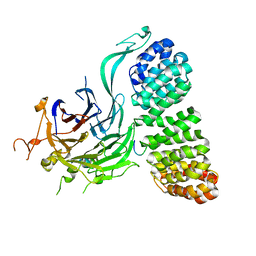 | | Cryo-EM structure of the PP2A:B55-FAM122A complex, B55 body | | Descriptor: | PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
8TWI
 
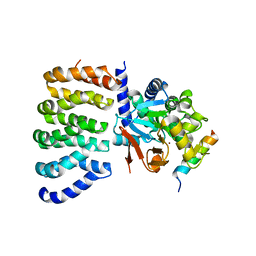 | | Cryo-EM structure of the PP2A:B55-FAM122A complex, PP2Ac body | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-08-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
8TTB
 
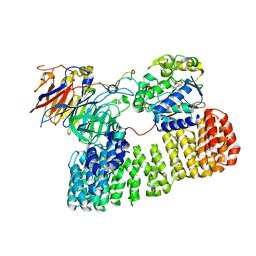 | | Cryo-EM structure of the PP2A:B55-ARPP19 complex | | Descriptor: | FE (III) ION, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-08-13 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
3Q86
 
 | |
3QCQ
 
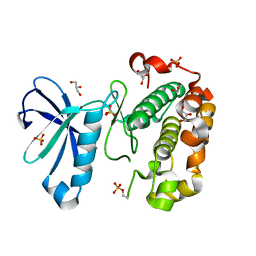 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-(3-Amino-1H-indazol-6-yl)-N4-ethyl-2,4-pyrimidinediamine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-(3-amino-2H-indazol-6-yl)-N~4~-ethylpyrimidine-2,4-diamine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3MQT
 
 | |
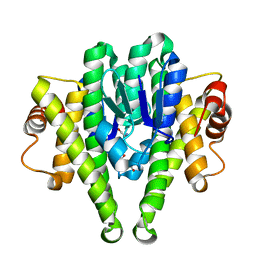
3M0F
 
 | |
3M7V
 
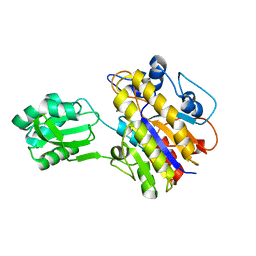 | | Crystal structure of phosphopentomutase from streptococcus mutans | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Fedorov, A.A, Bonanno, J, Fedorov, E.V, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphopentomutase from streptococcus mutans
To be Published
|
|
3M8N
 
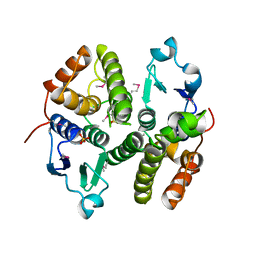 | |
3R95
 
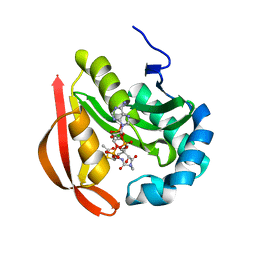 | |
3MDK
 
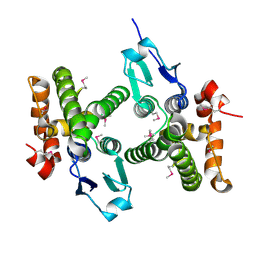 | |
3R9F
 
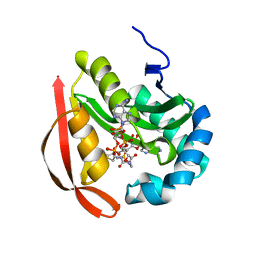 | |
3MC1
 
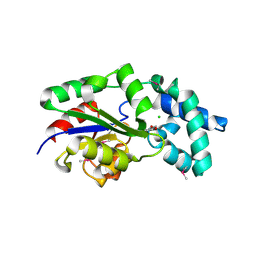 | | Crystal structure of a predicted phosphatase from Clostridium acetobutylicum | | Descriptor: | CHLORIDE ION, GLYCEROL, Predicted phosphatase, ... | | Authors: | Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-26 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of a predicted phosphatase from Clostridium acetobutylicum
To be Published, 2010
|
|
3M2P
 
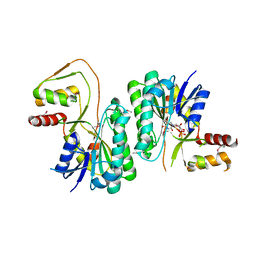 | |
3F48
 
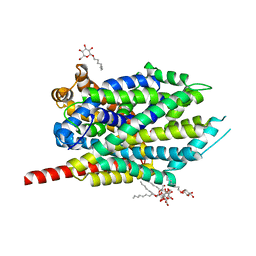 | | Crystal structure of LeuT bound to L-alanine and sodium | | Descriptor: | ALANINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-31 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3R9G
 
 | |
3QD3
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl {(3R,6S)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3MJE
 
 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | Descriptor: | AmphB, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | Deposit date: | 2010-04-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
3MSY
 
 | |
3M3P
 
 | | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus | | Descriptor: | Glutamine amido transferase | | Authors: | Fedorov, A.A, Domagalski, M, Fedorov, E.V, Burley, S.K, Minor, W, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus
To be Published
|
|
3QKC
 
 | | CRYSTAL STRUCTURE OF geranyl diphosphate synthase small subunit from Antirrhinum majus | | Descriptor: | Geranyl diphosphate synthase small subunit | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of geranyl diphosphate synthase small subunit from Antirrhinum majus
To be Published
|
|
