3KKE
 
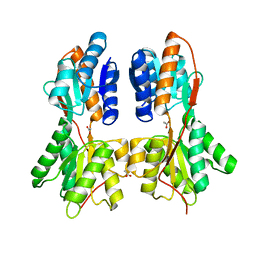 | | Crystal structure of a LacI family transcriptional regulator from Mycobacterium smegmatis | | Descriptor: | ACETATE ION, LacI family Transcriptional regulator | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Do, J, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a LacI family transcriptional regulator from Mycobacterium smegmatis
To be Published
|
|
2GHK
 
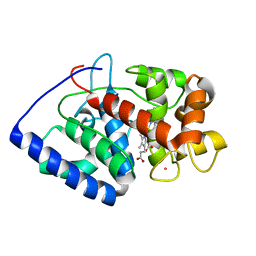 | | Conformational mobility in the active site of a heme peroxidase | | Descriptor: | CYANIDE ION, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Badyal, S.K, Joyce, M.G, Sharp, K.H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Conformational Mobility in the Active Site of a Heme Peroxidase.
J.Biol.Chem., 281, 2006
|
|
3KCM
 
 | |
3KXW
 
 | |
3KGZ
 
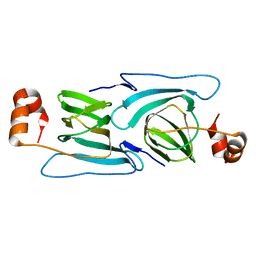 | | Crystal structure of a cupin 2 conserved barrel domain protein from Rhodopseudomonas palustris | | Descriptor: | Cupin 2 conserved barrel domain protein, MANGANESE (II) ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-29 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a cupin 2 conserved barrel domain protein from Rhodopseudomonas palustris
To be Published
|
|
2G02
 
 | | Nisin cyclase | | Descriptor: | Nisin biosynthesis protein nisC, ZINC ION | | Authors: | Nair, S.K. | | Deposit date: | 2006-02-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of the Lantibiotic Cyclase Involved in Nisin Biosynthesis
Science, 311, 2006
|
|
3KHN
 
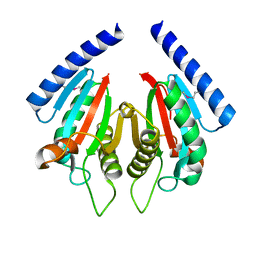 | |
3KHT
 
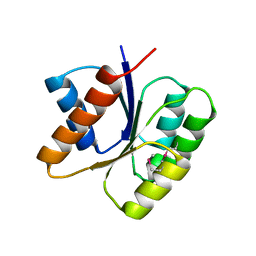 | |
7M74
 
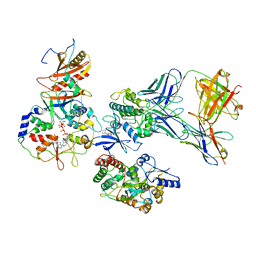 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
2G9K
 
 | | Human Transthyretin (TTR) Complexed with Hydroxylated polychlorinated Biphenyl-4-hydroxy-2',3,3',4',5-Pentachlorobiphenyl | | Descriptor: | 2',3,3',4',5-PENTACHLOROBIPHENYL-4-OL, Transthyretin | | Authors: | Palaninathan, S.K, Smith, C, Safe, S.H, Kelly, J.W, Sacchettini, J.C. | | Deposit date: | 2006-03-06 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hydroxylated polychlorinated biphenyls selectively bind transthyretin in blood and inhibit amyloidogenesis: rationalizing rodent PCB toxicity
Chem.Biol., 11, 2004
|
|
6USM
 
 | |
3KJX
 
 | |
3KES
 
 | | Crystal structure of the autoproteolytic domain from the nuclear pore complex component NUP145 from Saccharomyces cerevisiae in the Hexagonal, P61 space group | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin NUP145 | | Authors: | Sampathkumar, P, Ozyurt, S.A, Do, J, Bain, K, Dickey, M, Gheyi, T, Sali, A, Kim, S.J, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Rout, M, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Proteins, 78, 2010
|
|
2HDF
 
 | |
4RIM
 
 | |
7MSK
 
 | | ThuS glycosin S-glycosyltransferase | | Descriptor: | Glyco_trans_2-like domain-containing protein, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
7MSP
 
 | | SunS glycosin S-glycosyltransferase | | Descriptor: | MAGNESIUM ION, SPbeta prophage-derived glycosyltransferase SunS, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
4RIP
 
 | |
3KD6
 
 | |
7MSN
 
 | | SunS glycosin S-glycosyltransferase | | Descriptor: | SPbeta prophage-derived glycosyltransferase SunS, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
3KQ5
 
 | | Crystal structure of an uncharacterized protein from Coxiella burnetii | | Descriptor: | Hypothetical cytosolic protein | | Authors: | Bonanno, J.B, Freeman, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-17 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an uncharacterized protein from Coxiella burnetii
To be Published
|
|
6UVQ
 
 | | Crystal structure of Apo AtmM | | Descriptor: | ACETATE ION, D-glucose O-methyltransferase, MAGNESIUM ION | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Apo AtmM
To Be Published
|
|
3KRT
 
 | | CRYSTAL STRUCTURE OF putative crotonyl CoA reductase from Streptomyces coelicolor A3(2) | | Descriptor: | CHLORIDE ION, Crotonyl CoA reductase | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-19 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | CRYSTAL STRUCTURE OF putative crotonyl CoA reductase from Streptomyces coelicolor A3(2)
To be Published
|
|
3KSM
 
 | | Crystal structure of ABC-type sugar transport system, periplasmic component from Hahella chejuensis | | Descriptor: | ABC-type sugar transport system, periplasmic component, beta-D-ribofuranose | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-15 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ABC-type sugar transport system, periplasmic component from Hahella chejuensis
To be Published
|
|
3KSU
 
 | | Crystal structure of short-chain dehydrogenase from oenococcus oeni psu-1 | | Descriptor: | 3-oxoacyl-acyl carrier protein reductase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase from Oenococcus Oeni Psu-1
To be Published
|
|
