3NAX
 
 | | PDK1 in complex with inhibitor MP7 | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and pharmacological inhibition of PDK1 in cancer cells: characterization of a selective allosteric kinase inhibitor.
J.Biol.Chem., 286, 2011
|
|
3NEH
 
 | | Crystal structure of the protein LMO2462 from Listeria monocytogenes complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Renal dipeptidase family protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of the protein LMO2462 from Listeria monocytogenes
complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala
To be Published
|
|
3N8N
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 6 | | Descriptor: | (1R,4R,5R)-3-(tert-butylcarbamoyl)-1,4,5-trihydroxycyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3N7A
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 2 | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3N8K
 
 | | Type II dehydroquinase from Mycobacterium tuberculosis complexed with citrazinic acid | | Descriptor: | 2,6-dioxo-1,2,3,6-tetrahydropyridine-4-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Snee, W.C, Palaninathan, S.K, Sacchettini, J.C, Dias, M.V.B, Bromfield, K.M, Payne, R, Ciulli, A, Howard, N.I, Abell, C, Blundell, T.L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3NAY
 
 | | PDK1 in complex with inhibitor MP6 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 4-(2-cyclopropylethylidene)-9-(1H-pyrazol-4-yl)-6-{[(1R)-1,2,2-trimethylpropyl]amino}benzo[c][1,6]naphthyridin-1(4H)-one | | Authors: | Yan, Y, Munshi, S.K, Allison, T. | | Deposit date: | 2010-06-02 | | Release date: | 2010-11-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective inhibition of PDK1 using an allosteric kinase inhibitor and RNAi impairs cancer cell migration and anchorage-independent growth of primary tumor lines
J.Biol.Chem., 2010
|
|
3QDK
 
 | |
3QCS
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-[2-Amino-6-(4-morpholinyl)-4-pyrimidinyl]-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-[2-amino-6-(morpholin-4-yl)pyrimidin-4-yl]-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3NIW
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Bonanno, J.B, Ramagopal, U, Toro, R, Rutter, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-16 | | Release date: | 2010-06-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron
To be Published
|
|
3QLD
 
 | |
1B54
 
 | | CRYSTAL STRUCTURE OF A YEAST HYPOTHETICAL PROTEIN-A STRUCTURE FROM BNL'S HUMAN PROTEOME PROJECT | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, YEAST HYPOTHETICAL PROTEIN | | Authors: | Swaminathan, S, Eswaramoorthy, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-01-12 | | Release date: | 1999-01-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3LYB
 
 | |
3Q89
 
 | |
3Q8U
 
 | |
3LYP
 
 | |
3QD0
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with (2R,5S)-1-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-N-phenyl-3-piperidinecarboxamide | | Descriptor: | (3S,6R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-6-methyl-N-phenylpiperidine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QFW
 
 | | Crystal structure of Rubisco-like protein from Rhodopseudomonas palustris | | Descriptor: | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-01-23 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of Rubisco-like protein from Rhodopseudomonas palustris
To be Published
|
|
3ME7
 
 | | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5 | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative electron transport protein aq_2194 from Aquifex aeolicus VF5
To be Published
|
|
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3NO1
 
 | |
1AN2
 
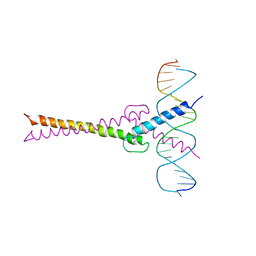 | | RECOGNITION BY MAX OF ITS COGNATE DNA THROUGH A DIMERIC B/HLH/Z DOMAIN | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*GP*GP*TP*CP*AP*CP*GP*TP*GP*AP*CP*C P*TP*AP*CP*AP*C)- 3'), PROTEIN (TRANSCRIPTION FACTOR MAX (TF MAX)) | | Authors: | Ferre-D'Amare, A.R, Prendergast, G.C, Ziff, E.B, Burley, S.K. | | Deposit date: | 1996-09-06 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain.
Nature, 363, 1993
|
|
5XJD
 
 | | TEAD in complex with fragment | | Descriptor: | (2S)-2-phenyl-2-pyrrol-1-yl-ethanoic acid, Transcriptional enhancer factor TEF-3 | | Authors: | Kaan, H.Y.K, Sim, A.Y.L, Tan, S.K.J, Verma, C, Song, H. | | Deposit date: | 2017-05-01 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Targeting YAP/TAZ-TEAD protein-protein interactions using fragment-based and computational modeling approaches.
PLoS ONE, 12, 2017
|
|
3NSF
 
 | | Apo form of the multicopper oxidase CueO | | Descriptor: | Blue copper oxidase cueO | | Authors: | Roberts, S.A, Montfort, W.R, Singh, S.K. | | Deposit date: | 2010-07-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
3NPK
 
 | |
3RAZ
 
 | |
