6C0H
 
 | |
6C9T
 
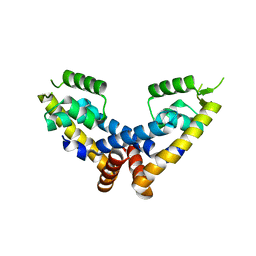 | | Transcriptional repressor, CouR | | Descriptor: | CouR | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-28 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6D6L
 
 | |
6C28
 
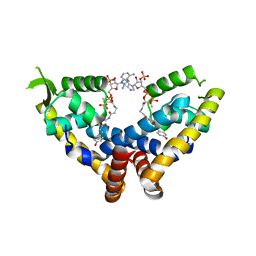 | | Transcriptional repressor, CouR, bound to p-coumaroyl-CoA | | Descriptor: | Transcriptional regulator, MarR family, p-coumaroyl-CoA | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-07 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6CGQ
 
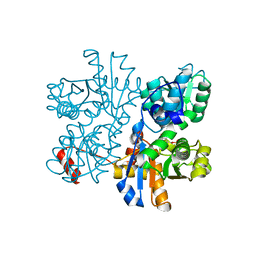 | | Threonine synthase from Bacillus subtilis ATCC 6633 with PLP and PLP-Ala | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Petronikolou, N, Nair, S.K. | | Deposit date: | 2018-02-20 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Molecular Basis of Bacillus subtilis ATCC 6633 Self-Resistance to the Phosphono-oligopeptide Antibiotic Rhizocticin.
ACS Chem. Biol., 14, 2019
|
|
6D6N
 
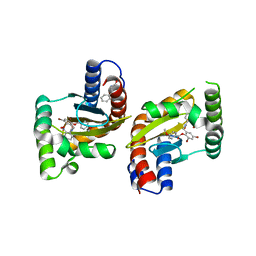 | | The structure of ligand binding domain of LasR in complex with TP-1 homolog, compound 16 | | Descriptor: | 2,4-dibromo-6-{[(2-nitrobenzene-1-carbonyl)amino]methyl}phenyl 4-methoxybenzoate, PHENYLALANINE, Transcriptional activator protein LasR | | Authors: | Dong, S.H, Nair, S.K. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Biochemical Studies of Non-native Agonists of the LasR Quorum-Sensing Receptor Reveal an L3 Loop "Out" Conformation for LasR.
Cell Chem Biol, 25, 2018
|
|
6C0G
 
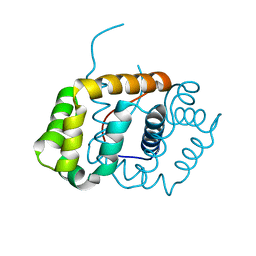 | |
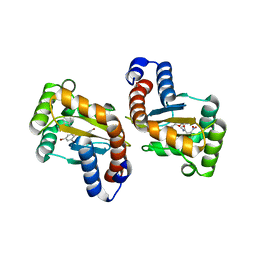
6D6M
 
 | |
6D6A
 
 | |
6C2S
 
 | | Transcriptional repressor, CouR, bound to a 23-mer DNA duplex | | Descriptor: | 23-mer, Transcriptional regulator, MarR family | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-08 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6C8R
 
 | |
6C0Y
 
 | |
6C8S
 
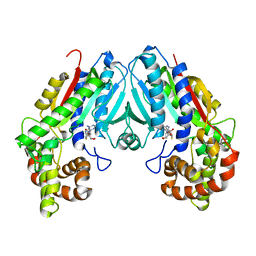 | | Loganic acid methyltransferase with SAH | | Descriptor: | Loganic acid O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Petronikolou, N, Nair, S.K. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Loganic Acid Methyltransferase: Insights into the Specificity of Methylation on an Iridoid Glycoside.
Chembiochem, 19, 2018
|
|
4IIY
 
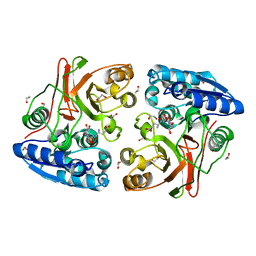 | |
2KNA
 
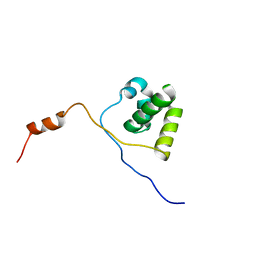 | | Solution structure of UBA domain of XIAP | | Descriptor: | Baculoviral IAP repeat-containing protein 4 | | Authors: | Hui, S.K, Tse, M.K, Sze, K.H. | | Deposit date: | 2009-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Backbone and side-chain 1H, 13C and 15N assignments of the ubiquitin-associated domain of human X-linked inhibitor of apoptosis protein
Biomol.Nmr Assign., 4, 2010
|
|
2K8R
 
 | |
4IL2
 
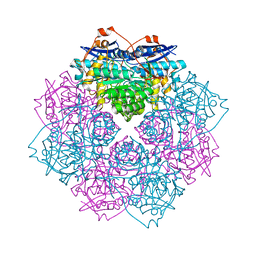 | | Crystal structure of D-mannonate dehydratase (rspA) from E. coli CFT073 (EFI TARGET EFI-501585) | | Descriptor: | MAGNESIUM ION, Starvation sensing protein rspA | | Authors: | Lukk, T, Wichelecki, D, Imker, H.J, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-12-28 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mannonate degradation pathway in E. coli CFT073
To be Published
|
|
2K8M
 
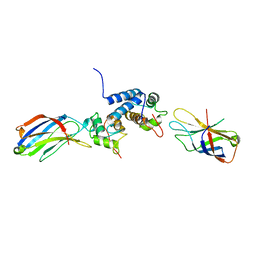 | | S100A13-C2A binary complex structure | | Descriptor: | Protein S100-A13, Putative uncharacterized protein | | Authors: | Mohan, S.K, Rani, S.G, Kumar, S.M, Yu, C. | | Deposit date: | 2008-09-14 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | S100A13-C2A binary complex structure-a key component in the acidic fibroblast growth factor for the non-classical pathway.
Biochem.Biophys.Res.Commun., 380, 2009
|
|
2LR4
 
 | |
4II2
 
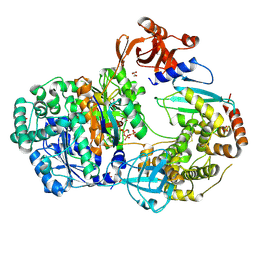 | | Crystal structure of Ubiquitin activating enzyme 1 (Uba1) in complex with the Ub E2 Ubc4, ubiquitin, and ATP/Mg | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Olsen, S.K, Lima, C.D. | | Deposit date: | 2012-12-19 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer.
Mol.Cell, 49, 2013
|
|
4IKH
 
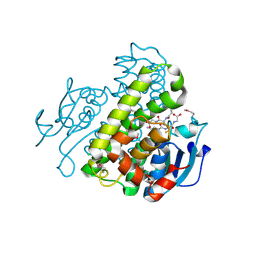 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-26 | | Release date: | 2013-01-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound
To be Published
|
|
4II3
 
 | |
4IIX
 
 | |
4IL0
 
 | | Crystal structure of GlucDRP from E. coli K-12 MG1655 (EFI target EFI-506058) | | Descriptor: | CITRIC ACID, GLYCEROL, Glucarate dehydratase-related protein | | Authors: | Lukk, T, Ghasempur, S, Imker, H.J, Gerlt, J.A, Nair, S.K, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-28 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Glucarate dehydratase and its related protein from Escherichia coli form a heterotetrameric complex.
to be published
|
|
4IJI
 
 | | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione | | Descriptor: | ACRYLIC ACID, BENZOIC ACID, Glutathione S-transferase-like protein YibF, ... | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Psuedomonas fluorescens Pf-5, target EFI-900011, with bound S-(propanoic acid)-glutathione
To be Published
|
|
