3OID
 
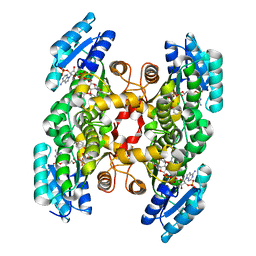 | | Crystal Structure of Enoyl-ACP Reductases III (FabL) from B. subtilis (complex with NADP and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3N3D
 
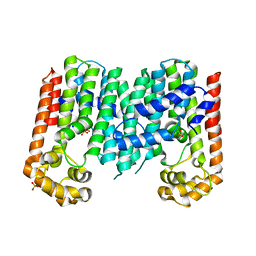 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Geranylgeranyl pyrophosphate synthase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis
To be Published
|
|
2NW9
 
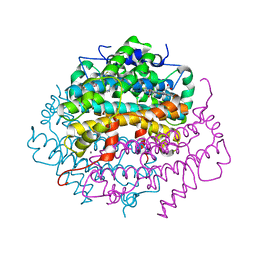 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferrous heme and 6-fluoro-tryptophan. Northeast Structural Genomics Target XcR13 | | Descriptor: | 6-FLUORO-L-TRYPTOPHAN, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Bruckmann, C, Thackray, S.J, Seetharaman, J, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3N6J
 
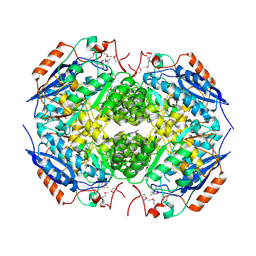 | | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus succinogenes 130Z | | Descriptor: | Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-25 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus
succinogenes 130Z
To be Published
|
|
3OIF
 
 | | Crystal Structure of Enoyl-ACP Reductases I (FabI) from B. subtilis (complex with NAD and TCL) | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kim, K.-H, Ha, B.H, Kim, S.J, Hong, S.K, Hwang, K.Y, Kim, E.E. | | Deposit date: | 2010-08-19 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Enoyl-ACP Reductases I (FabI) and III (FabL) from B. subtilis
J.Mol.Biol., 406, 2011
|
|
3N87
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 3 | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[3-(phenylcarbonyl)phenyl]cyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
1YD9
 
 | | 1.6A Crystal Structure of the Non-Histone Domain of the Histone Variant MacroH2A1.1. | | Descriptor: | Core histone macro-H2A.1, GOLD ION | | Authors: | Chakravarthy, S, Swamy, G.Y.S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the histone variant macroH2A
Mol.Cell.Biol., 25, 2005
|
|
3NEK
 
 | | Crystal structure of a nitrogen repressor-like protein MJ0159 from Methanococcus jannaschii | | Descriptor: | GLYCEROL, nitrogen repressor-like protein MJ0159 | | Authors: | Bonanno, J.B, Patskovsky, Y, Malashkevich, V, Ozyurt, S, Dickey, M, Wu, B, Maletic, M, Rodgers, L, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural underpinnings of nitrogen regulation by the prototypical nitrogen-responsive transcriptional factor NrpR.
Structure, 18, 2010
|
|
3N6H
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus succinogenes 130Z complexed with magnesium/sulfate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-25 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus
succinogenes 130Z complexed with magnesium/sulfate
To be Published
|
|
3F4I
 
 | | Crystal Structure of LeuT bound to L-selenomethionine and sodium | | Descriptor: | SELENOMETHIONINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-31 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
8TEK
 
 | | Baseplate of Nexin-dynein regulatory complex from Tetrahymena thermophila | | Descriptor: | CFAP20, Cilia- and flagella-associated protein 91, Coiled-coil protein, ... | | Authors: | Ghanaeian, A.G, Black, C.S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Integrated modeling of the Nexin-dynein regulatory complex reveals its regulatory mechanism.
Nat Commun, 14, 2023
|
|
8TID
 
 | | Combined linker domain of N-DRC and associated proteins Tetrahymena | | Descriptor: | AAA family ATPase CDC48 subfamily protein, CFAP20, Calmodulin 7-2, ... | | Authors: | Ghanaeian, A.G, Majhi, S.M, McCaffrey, C.M, Nami, B.N, Black, C.B, Yang, S.K, Legal, T.L, Papoulas, O.P, Janowska, M.J, Valente-Paterno, M.V, Marcotte, E.M, Wloga, D.W, Bui, K.H. | | Deposit date: | 2023-07-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Integrated modeling of the Nexin-dynein regulatory complex reveals its regulatory mechanism.
Nat Commun, 14, 2023
|
|
3PLU
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDI) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
3M1Y
 
 | |
3MIZ
 
 | |
3RIT
 
 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg and dipeptide L-Arg-D-Lys | | Descriptor: | ARGININE, D-LYSINE, Dipeptide epimerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-14 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MN1
 
 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a | | Descriptor: | CHLORIDE ION, probable yrbi family phosphatase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3R96
 
 | |
3MPO
 
 | |
3MSR
 
 | |
3IK4
 
 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Herpetosiphon aurantiacus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing protein, POTASSIUM ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Iizuka, M, Sauder, J.M, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3KL2
 
 | | Crystal structure of a putative isochorismatase from Streptomyces avermitilis | | Descriptor: | Putative isochorismatase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative isochorismatase from Streptomyces avermitilis
To be Published
|
|
8SLB
 
 | | X-ray structure of CorA N-terminal domain in complex with conformation-specific synthetic antibody C12 | | Descriptor: | CHLORIDE ION, Cobalt/magnesium transport protein CorA, sAB C12 Heavy Chain, ... | | Authors: | Dominik, P.K, Erramilli, S.K, Reddy, B.G, Kossiakoff, A.A. | | Deposit date: | 2023-04-21 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Conformation-specific Synthetic Antibodies Discriminate Multiple Functional States of the Ion Channel CorA.
J.Mol.Biol., 435, 2023
|
|
3KQ4
 
 | | Structure of Intrinsic Factor-Cobalamin bound to its receptor Cubilin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Andersen, C.B.F, Madsen, M, Moestrup, S.K, Andersen, G.R. | | Deposit date: | 2009-11-17 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes.
Nature, 464, 2010
|
|
3KTS
 
 | | CRYSTAL STRUCTURE OF GLYCEROL UPTAKE OPERON ANTITERMINATOR REGULATORY PROTEIN FROM LISTERIA MONOCYTOGENES STR. 4b F2365 | | Descriptor: | Glycerol uptake operon antiterminator regulatory protein, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | CRYSTAL STRUCTURE OF GLYCEROL UPTAKE OPERON ANTITERMINATOR REGULATORY PROTEIN FROM LISTERIA MONOCYTOGENES STR. 4b F2365
To be Published
|
|
