5Q0L
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-{2-[4-(hydroxymethyl)phenyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0Y
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-{5,6-difluoro-2-[6-(1H-pyrazol-1-yl)pyridin-3-yl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1F
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5-fluoro-1H-benzimidazol-1-yl]acetyl}amino)cyclohexane-1-carboxylic acid | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0S
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(2-cyanophenyl)-2-cyclohexylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q17
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 4-({5-bromo-2-oxo-1'-[(thiophen-2-yl)sulfonyl]spiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, Nuclear receptor coactivator peptide SRC2 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1H
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-N,2-dicyclohexyl-2-{2-[4-(1H-tetrazol-5-yl)phenyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0N
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 3-chloro-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q10
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N-[3-(acetylamino)phenyl]-4-chloro-N-[(1S)-1-cyclohexyl-2-(cyclohexylamino)-2-oxoethyl]benzamide | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1D
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexyl-N-phenylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
1MWA
 
 | | 2C/H-2KBM3/DEV8 ALLOGENEIC COMPLEX | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2C T CELL RECEPTOR ALPHA CHAIN, ... | | 著者 | Luz, J.G, Huang, M.D, Garcia, K.C, Rudolph, M.G, Teyton, L, Wilson, I.A. | | 登録日 | 2002-09-27 | | 公開日 | 2002-11-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural comparison of allogeneic and syngeneic T cell receptor-peptide-major histocompatibility
complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially
increasing V(beta) Interactions.
J.EXP.MED., 195, 2002
|
|
8OFB
 
 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch, hexagonal form | | 分子名称: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Reverse gyrase, ... | | 著者 | Klostermeier, D, Rasche, R, Mhaindarkar, V, Kummel, D, Rudolph, M.G. | | 登録日 | 2023-03-15 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7FVS
 
 | | Crystal Structure of S. aureus gyrase in complex with 4-[[1-[(1-chloro-6,7-dihydro-5H-cyclopenta[c]pyridin-6-yl)methyl]azetidin-3-yl]methylamino]-6-fluorochromen-2-one | | 分子名称: | 4-{[(1-{[(6R)-1-chloro-6,7-dihydro-5H-cyclopenta[c]pyridin-6-yl]methyl}azetidin-3-yl)methyl]amino}-6-fluoro-2H-1-benzopyran-2-one, ACETATE ION, CHLORIDE ION, ... | | 著者 | Xu, B, Benz, J, Cumming, J.G, Kreis, L, Rudolph, M.G. | | 登録日 | 2023-04-18 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Discovery of a Series of Indane-Containing NBTIs with Activity against Multidrug-Resistant Gram-Negative Pathogens.
Acs Med.Chem.Lett., 14, 2023
|
|
7FVT
 
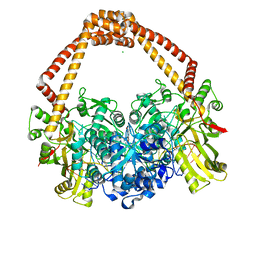 | | Crystal Structure of S. aureus gyrase in complex with 6-[5-[2-[(4-chloro-2,3-dihydro-1H-inden-2-yl)methylamino]ethyl]-2-oxo-1,3-oxazolidin-3-yl]-4H-pyrido[3,2-b][1,4]oxazin-3-one | | 分子名称: | 6-{(5R)-5-[2-({[(2R)-4-chloro-2,3-dihydro-1H-inden-2-yl]methyl}amino)ethyl]-2-oxo-1,3-oxazolidin-3-yl}-2H-pyrido[3,2-b][1,4]oxazin-3(4H)-one, CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Xu, B, Benz, J, Cumming, J.G, Rudolph, M.G. | | 登録日 | 2023-04-18 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.081 Å) | | 主引用文献 | Discovery of a Series of Indane-Containing NBTIs with Activity against Multidrug-Resistant Gram-Negative Pathogens.
Acs Med.Chem.Lett., 14, 2023
|
|
8S1K
 
 | | Crystal Structure of human FABP4 in complex with 2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1H-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline | | 分子名称: | (6~{S})-2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst-Sander, U, Rudolph, M.G. | | 登録日 | 2024-02-15 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Crystal Structure of a human FABP4 complex
To be published
|
|
5O0V
 
 | |
5LSA
 
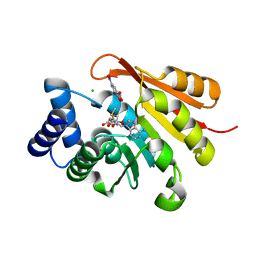 | |
5LQJ
 
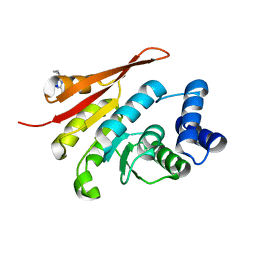 | | Crystal Structure of COMT in complex with 3-cyclopropyl-5-methyl-4-phenyl-1,2,4-triazole | | 分子名称: | 3-cyclopropyl-5-methyl-4-phenyl-1,2,4-triazole, CHLORIDE ION, Catechol O-methyltransferase, ... | | 著者 | Ehler, A, Lerner, C, Rudolph, M.G. | | 登録日 | 2016-08-17 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal Structure of COMT in complex with 3-cyclopropyl-5-methyl-4-phenyl-1,2,4-triazole
To be published
|
|
5LR6
 
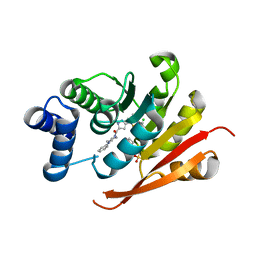 | | Crystal Structure of COMT in complex with [3-(2,4-dimethyl-1,3-thiazol-5-yl)-1H-pyrazol-5-yl]-(4-phenylpiperazin-1-yl)methanone | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | 著者 | Ehler, A, Lerner, C, Rudolph, M.G. | | 登録日 | 2016-08-18 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of COMT in complex with [3-(2,4-dimethyl-1,3-thiazol-5-yl)-1H-pyrazol-5-yl]-(4-phenylpiperazin-1-yl)methanone
To be published
|
|
5M6Q
 
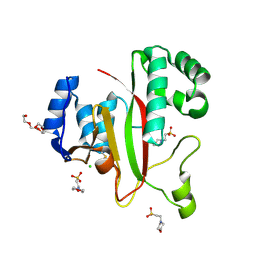 | | Crystal Structure of Kutzneria albida transglutaminase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Steffen, W, Benz, J, Rudolph, M.G. | | 登録日 | 2016-10-25 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Discovery of a microbial transglutaminase enabling highly site-specific labeling of proteins.
J. Biol. Chem., 292, 2017
|
|
6QZH
 
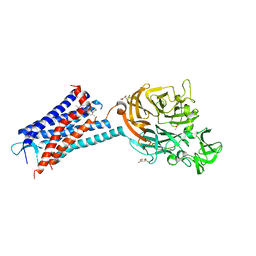 | | Structure of the human CC Chemokine Receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 and the insertion protein Sialidase NanA | | 分子名称: | 3-[[4-[[(1~{R})-2,2-dimethyl-1-(5-methylfuran-2-yl)propyl]amino]-1,1-bis(oxidanylidene)-1,2,5-thiadiazol-3-yl]amino]-~{N},~{N},6-trimethyl-2-oxidanyl-benzamide, C-C chemokine receptor type 7,Sialidase A,C-C chemokine receptor type 7, D(-)-TARTARIC ACID, ... | | 著者 | Jaeger, K, Bruenle, S, Weinert, T, Guba, W, Muehle, J, Miyazaki, T, Weber, M, Furrer, A, Haenggi, N, Tetaz, T, Huang, C.Y, Mattle, D, Vonach, J.M, Gast, A, Kuglstatter, A, Rudolph, M.G, Nogly, P, Benz, J, Dawson, R.J.P, Standfuss, J. | | 登録日 | 2019-03-11 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7.
Cell, 178, 2019
|
|
3U81
 
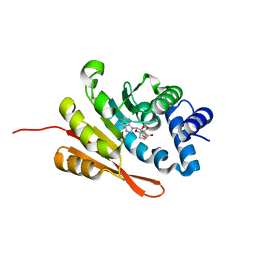 | | Crystal structure of a SAH-bound semi-holo form of rat Catechol-O-methyltransferase | | 分子名称: | Catechol O-methyltransferase, POTASSIUM ION, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | 登録日 | 2011-10-15 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Catechol-O-methyltransferase in complex with substituted 3'-deoxyribose bisubstrate inhibitors.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8ARU
 
 | |
1G6R
 
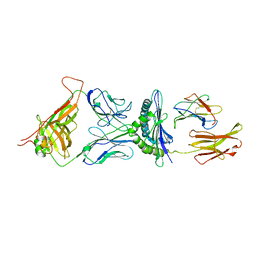 | | A FUNCTIONAL HOT SPOT FOR ANTIGEN RECOGNITION IN A SUPERAGONIST TCR/MHC COMPLEX | | 分子名称: | ALPHA T CELL RECEPTOR, BETA T CELL RECEPTOR, BETA-2 MICROGLOBULIN, ... | | 著者 | Degano, M, Garcia, K.C, Apostolopoulos, V, Rudolph, M.G, Teyton, L, Wilson, I.A. | | 登録日 | 2000-11-07 | | 公開日 | 2000-11-15 | | 最終更新日 | 2018-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A functional hot spot for antigen recognition in a superagonist TCR/MHC complex.
Immunity, 12, 2000
|
|
1HQ8
 
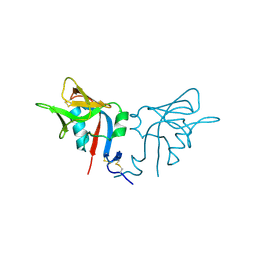 | | CRYSTAL STRUCTURE OF THE MURINE NK CELL-ACTIVATING RECEPTOR NKG2D AT 1.95 A | | 分子名称: | NKG2-D | | 著者 | Wolan, D.W, Teyton, L, Rudolph, M.G, Villmow, B, Bauer, S, Busch, D.H, Wilson, I.A. | | 登録日 | 2000-12-14 | | 公開日 | 2001-03-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A.
Nat.Immunol., 2, 2001
|
|
4JS1
 
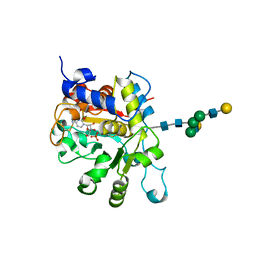 | | crystal structure of human Beta-galactoside alpha-2,6-sialyltransferase 1 in complex with cytidine and phosphate | | 分子名称: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Beta-galactoside alpha-2,6-sialyltransferase 1, PHOSPHATE ION, ... | | 著者 | Kuhn, B, Benz, J, Greif, M, Engel, A.M, Sobek, H, Rudolph, M.G. | | 登録日 | 2013-03-22 | | 公開日 | 2013-07-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | The structure of human alpha-2,6-sialyltransferase reveals the binding mode of complex glycans.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
