4F3R
 
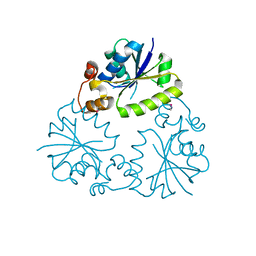 | | Structure of phosphopantetheine adenylyltransferase (CBU_0288) from Coxiella burnetii | | Descriptor: | CALCIUM ION, Phosphopantetheine adenylyltransferase | | Authors: | Franklin, M.C, Cheung, J, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-07-04 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
6FIL
 
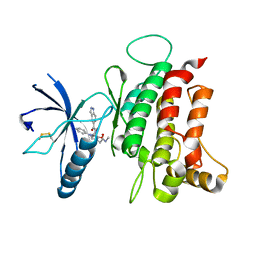 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.730A, P212121, Rfree=24.5% | | Descriptor: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FEX
 
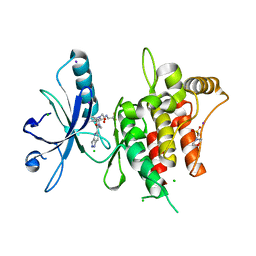 | | DDR1, 2-[4-bromo-2-oxo-1'-(1H-pyrazolo[4,3-b]pyridine-5-carbonyl)spiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide, 1.291A, P212121, Rfree=17.4% | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[4-bromanyl-2-oxidanylidene-1'-(1~{H}-pyrazolo[4,3-b]pyridin-5-ylcarbonyl)spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, CHLORIDE ION, ... | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
3HHD
 
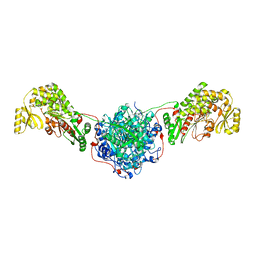 | | Structure of the Human Fatty Acid Synthase KS-MAT Didomain as a Framework for Inhibitor Design. | | Descriptor: | CHLORIDE ION, Fatty acid synthase | | Authors: | Pappenberger, G.M, Benz, J, Thoma, R, Rudolph, M.G. | | Deposit date: | 2009-05-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the human fatty acid synthase KS-MAT didomain as a framework for inhibitor design.
J.Mol.Biol., 397, 2010
|
|
6FIN
 
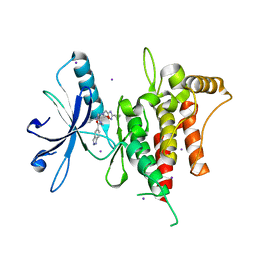 | | DDR1, 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1H-indazole-5-carbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, 1.670A, P1211, Rfree=22.8% | | Descriptor: | 3-[(3-cyclopropyl-1,2,4-oxadiazol-5-yl)methyl]-8-(1~{H}-indazol-5-ylcarbonyl)-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FIQ
 
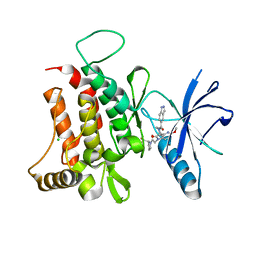 | | DDR1, 1-(1H-indazole-5-carbonyl)-5'-methoxy-1'-[2-oxo-2-[(2S)-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, 1.790A, P212121, Rfree=23.8% | | Descriptor: | 1-(1~{H}-indazol-5-ylcarbonyl)-5'-methoxy-1'-[2-oxidanylidene-2-[(2~{S})-2-(trifluoromethyl)pyrrolidin-1-yl]ethyl]spiro[piperidine-4,3'-pyrrolo[3,2-b]pyridine]-2'-one, CHLORIDE ION, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FEW
 
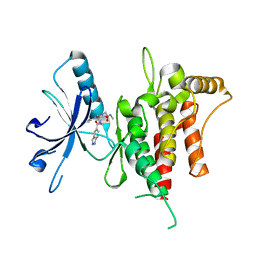 | | DDR1, 2-[8-(1H-indazole-5-carbonyl)-4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-N-methylacetamide, 1.440A, P1211, Rfree=24.1% | | Descriptor: | 2-[8-(2~{H}-indazol-5-ylcarbonyl)-4-oxidanylidene-1-phenyl-1,3,8-triazaspiro[4.5]decan-3-yl]-~{N}-methyl-ethanamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
6FIO
 
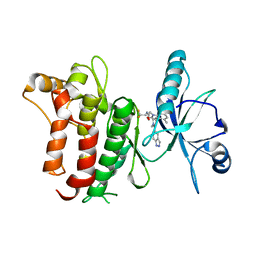 | | DDR1, 2-[1'-(1H-indazole-5-carbonyl)-4-methyl-2-oxospiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide, 1.990A, P6522, Rfree=27.7% | | Descriptor: | 2-[1'-(1~{H}-indazol-5-ylcarbonyl)-4-methyl-2-oxidanylidene-spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
5K03
 
 | | Crystal Structure of COMT in complex with 2,6-dimethyl-3-(1H-pyrazol-3-yl)imidazo[1,2-a]pyridine | | Descriptor: | 2,6-dimethyl-3-(1H-pyrazol-5-yl)imidazo[1,2-a]pyridine, Catechol O-methyltransferase, POTASSIUM ION | | Authors: | Ehler, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
5K0J
 
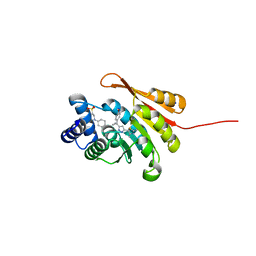 | | Crystal Structure of COMT in complex with 5-[5-[1-(4-methoxyphenyl)cyclopropyl]-1H-pyrazol-3-yl]-2,4-dimethyl-1,3-thiazole | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-{3-[1-(4-methoxyphenyl)cyclopropyl]-1H-pyrazol-5-yl}-2,4-dimethyl-1,3-thiazole, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
5K0G
 
 | | Crystal Structure of COMT in complex with 4-[5-[1-(4-methoxyphenyl)ethyl]-1H-pyrazol-3-yl]-1,3-dimethylpyrazole | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-[(1S)-1-(4-methoxyphenyl)ethyl]-1',3'-dimethyl-1'H,2H-3,4'-bipyrazole, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
5K0F
 
 | | Crystal Structure of COMT in complex with 5-[5-[1-(4-methoxyphenyl)ethyl]-1H-pyrazol-3-yl]-2,4-dimethyl-1,3-thiazole | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 5-{3-[(1R)-1-(4-methoxyphenyl)ethyl]-1H-pyrazol-5-yl}-2,4-dimethyl-1,3-thiazole, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Rodriguez-Sarmiento, R.M, Rudolph, M.G. | | Deposit date: | 2016-05-17 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Design of Potent and Druglike Nonphenolic Inhibitors for Catechol O-Methyltransferase Derived from a Fragment Screening Approach Targeting the S-Adenosyl-l-methionine Pocket.
J. Med. Chem., 59, 2016
|
|
3L0K
 
 | |
3L0N
 
 | | Human orotidyl-5'-monophosphate decarboxylase in complex with 6-mercapto-UMP | | Descriptor: | 6-sulfanyluridine-5'-phosphate, Uridine 5'-monophosphate synthase | | Authors: | Heinrich, D, Wittmann, J, Diederichsen, U, Rudolph, M. | | Deposit date: | 2009-12-10 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
4EY8
 
 | | Crystal structure of recombinant human acetylcholinesterase in complex with fasciculin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, Fasciculin-2, ... | | Authors: | Cheung, J, Rudolph, M, Burshteyn, F, Cassidy, M, Gary, E, Love, J, Height, J, Franklin, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5958 Å) | | Cite: | Structures of human acetylcholinesterase in complex with pharmacologically important ligands.
J.Med.Chem., 55, 2012
|
|
4JS1
 
 | | crystal structure of human Beta-galactoside alpha-2,6-sialyltransferase 1 in complex with cytidine and phosphate | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Beta-galactoside alpha-2,6-sialyltransferase 1, PHOSPHATE ION, ... | | Authors: | Kuhn, B, Benz, J, Greif, M, Engel, A.M, Sobek, H, Rudolph, M.G. | | Deposit date: | 2013-03-22 | | Release date: | 2013-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The structure of human alpha-2,6-sialyltransferase reveals the binding mode of complex glycans.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3EWZ
 
 | |
4JS2
 
 | | Crystal structure of human Beta-galactoside alpha-2,6-sialyltransferase 1 in complex with CMP | | Descriptor: | Beta-galactoside alpha-2,6-sialyltransferase 1, CYTIDINE-5'-MONOPHOSPHATE, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuhn, B, Benz, J, Greif, M, Engel, A.M, Sobek, H, Rudolph, M.G. | | Deposit date: | 2013-03-22 | | Release date: | 2013-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of human alpha-2,6-sialyltransferase reveals the binding mode of complex glycans.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3HVH
 
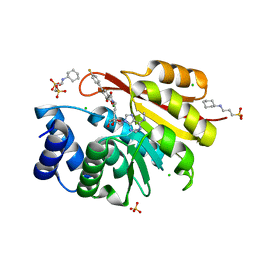 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-methyladenine-containing bisubstrate inhibitor | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HVI
 
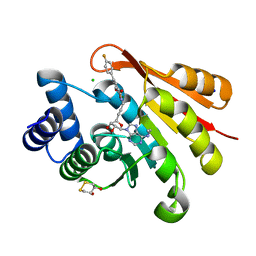 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-ethyladenine-containing bisubstrate inhibitor | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HVJ
 
 | | Rat catechol O-methyltransferase in complex with a catechol-type, N6-propyladenine-containing bisubstrate inhibitor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3HVK
 
 | | Rat catechol O-methyltransferase in complex with a catechol-type, purine-containing bisubstrate inhibitor - humanized form | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2009-06-16 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4NG4
 
 | | Structure of phosphoglycerate kinase (CBU_1782) from Coxiella burnetii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoglycerate kinase | | Authors: | Franklin, M.C, Cheung, J, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2013-11-01 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2QCF
 
 | |
2QCE
 
 | | Crystal structure of the orotidine-5'-monophosphate decarboxylase domain of human UMP synthase bound to sulfate, glycerol, and chloride | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Wittmann, J, Rudolph, M. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structures of the human orotidine-5'-monophosphate decarboxylase support a covalent mechanism and provide a framework for drug design.
Structure, 16, 2008
|
|
