4J2N
 
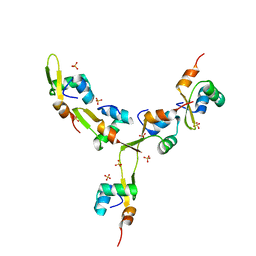 | | Crystal Structure of mycobacteriophage Pukovnik Xis | | Descriptor: | Gp37, SULFATE ION | | Authors: | Homa, N.J, Amrich, C.G, Heroux, A, VanDemark, A.P. | | Deposit date: | 2013-02-04 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | The Structure of Xis Reveals the Basis for Filament Formation and Insight into DNA Bending within a Mycobacteriophage Intasome.
J.Mol.Biol., 426, 2014
|
|
4GDX
 
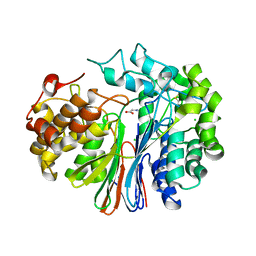 | | Crystal Structure of Human Gamma-Glutamyl Transpeptidase--Glutamate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
4L1P
 
 | |
4PXL
 
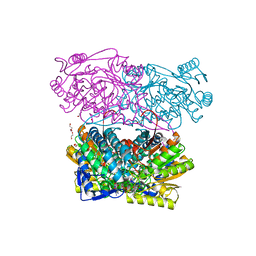 | | Structure of Zm ALDH2-3 (RF2C) in complex with NAD | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytosolic aldehyde dehydrogenase RF2C, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2014-03-24 | | Release date: | 2015-03-18 | | Last modified: | 2015-05-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7.
Biochem.J., 468, 2015
|
|
3P1H
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23K/I92A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, thermonuclease | | Authors: | Clark, I.A, Caro, J.A, Sue, G, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2010-09-30 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Pressure unfolding effects of artificial cavities in proteins.
To be Published
|
|
4OAL
 
 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor CPPU in alternative spacegroup | | Descriptor: | 1-(2-chloropyridin-4-yl)-3-phenylurea, Cytokinin dehydrogenase 4, DIMETHYL SULFOXIDE, ... | | Authors: | Kopecny, D, Morera, S, Vigouroux, A, Koncitikova, R. | | Deposit date: | 2014-01-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3QB3
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92KL25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Sue, G, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-01-12 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Relocation of internal ionizable residues to cavities created by single alanine substitutions
To be Published
|
|
4EA5
 
 | | Structure of the glycoslyase domain of MBD4 bound to a 5hmU containing DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*(5HU)*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2012-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
3RZ5
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-pentyl-4-sulfamoylbenzamide, ... | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RYY
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, N-propyl-4-sulfamoylbenzamide, ZINC ION | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RZ8
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-hexyl-4-sulfamoylbenzamide, ... | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
4I8P
 
 | | Crystal structure of aminoaldehyde dehydrogenase 1a from Zea mays (ZmAMADH1a) | | Descriptor: | 1,2-ETHANEDIOL, Aminoaldehyde dehydrogenase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
3SK5
 
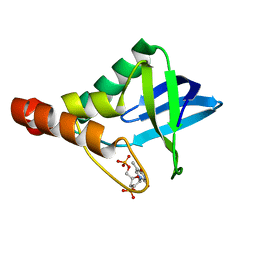 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V39D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V39D at cryogenic temperature
To be Published
|
|
4O95
 
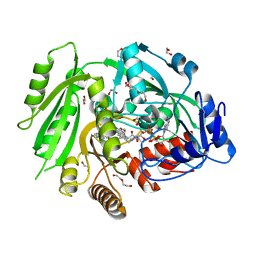 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor CPPU | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-chloropyridin-4-yl)-3-phenylurea, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Morera, S, Vigouroux, A, Koncitikova, R. | | Deposit date: | 2014-01-01 | | Release date: | 2015-04-01 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3OSO
 
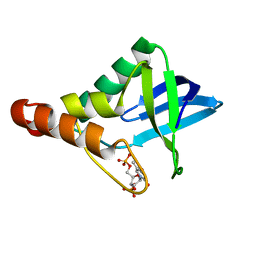 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OWF
 
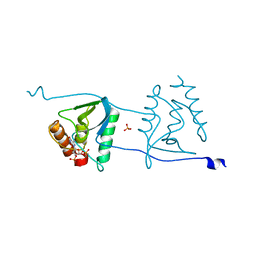 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66R at cryogenic temperature | | Descriptor: | CALCIUM ION, PHOSPHATE ION, THYMIDINE-3',5'-DIPHOSPHATE, ... | | Authors: | Schlessman, J.L, Khangulov, V, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2010-09-17 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Domain swapping promoted by a single mutation that introduces an ionizable group into the hydrophobic core of a protein
To be Published
|
|
4L1U
 
 | | Crystal Structure of Human Rtf1 Plus3 Domain in Complex with Spt5 CTR Phosphopeptide | | Descriptor: | GLYCEROL, RNA polymerase-associated protein RTF1 homolog, SULFATE ION, ... | | Authors: | Wier, A.D, Heroux, A, VanDemark, A.P. | | Deposit date: | 2013-06-03 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4PP0
 
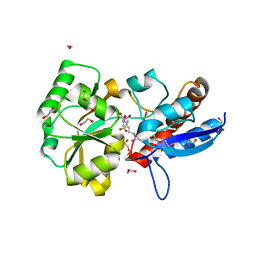 | | Structure of the PBP NocT-M117N in complex with pyronopaline | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1S)-4-carbamimidamido-1-carboxybutyl]-5-oxo-D-proline, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
3RLE
 
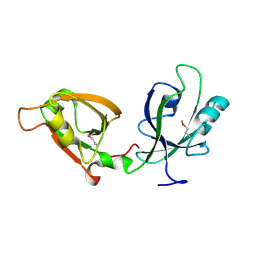 | | Crystal Structure of GRASP55 GRASP domain (residues 7-208) | | Descriptor: | Golgi reassembly-stacking protein 2 | | Authors: | Truschel, S.T, Sengupta, D, Foote, A, Heroux, A, Macbeth, M.R, Linstedt, A.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structure of the Membrane-tethering GRASP Domain Reveals a Unique PDZ Ligand Interaction That Mediates Golgi Biogenesis.
J.Biol.Chem., 286, 2011
|
|
4POD
 
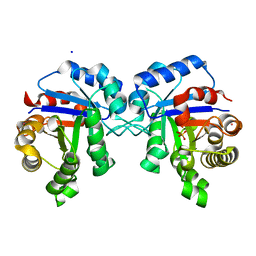 | | Structure of Triosephosphate Isomerase I170V mutant human enzyme. | | Descriptor: | BROMIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Amrich, C.G, Aslam, A.A, Heroux, A, VanDemark, A.P. | | Deposit date: | 2014-02-25 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
Biochim.Biophys.Acta, 1852, 2015
|
|
4POC
 
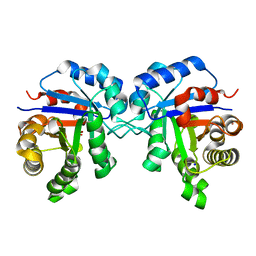 | | Structure of Triosephosphate Isomerase Wild Type human enzyme. | | Descriptor: | BROMIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Amrich, C.G, Aslam, A.A, Heroux, A, VanDemark, A.P. | | Deposit date: | 2014-02-25 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
Biochim.Biophys.Acta, 1852, 2015
|
|
3PMF
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2010-11-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4POW
 
 | | Structure of the PBP NocT in complex with pyronopaline | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1S)-4-carbamimidamido-1-carboxybutyl]-5-oxo-D-proline, Nopaline-binding periplasmic protein | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Agrobacterium uses a unique ligand-binding mode for trapping opines and acquiring a competitive advantage in the niche construction on plant host.
Plos Pathog., 10, 2014
|
|
3QOJ
 
 | | Cryogenic structure of Staphylococcal nuclease variant D+PHS/V23K | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2011-02-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determinants of pKa values of internal ionizable groups: properties of ion pairs in the protein core
To be Published
|
|
4I8Q
 
 | | Structure of the aminoaldehyde dehydrogenase 1 E260A mutant from Solanum lycopersicum (SlAMADH1-E260A) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2012-12-04 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Plant ALDH10 family: identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate.
J.Biol.Chem., 288, 2013
|
|
