2KJX
 
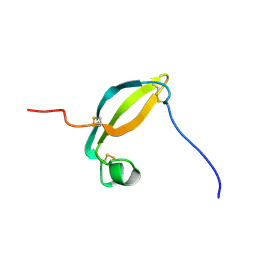 | | Solution structure of the extracellular domain of JTB | | Descriptor: | Jumping translocation breakpoint protein | | Authors: | Rousseau, F, Lingel, A, Pan, B, Fairbrother, W.J, Bazan, F. | | Deposit date: | 2009-06-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structure of the extracellular domain of the jumping translocation breakpoint protein reveals a variation of the midkine fold.
J.Mol.Biol., 415, 2012
|
|
4Z33
 
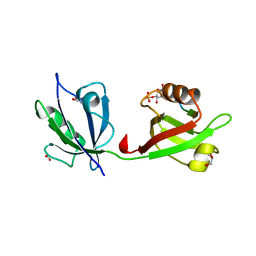 | | Crystal structure of the syntenin PDZ1 and PDZ2 tandem in complex with the Frizzled 7 C-terminal fragment and PIP2 | | Descriptor: | ACETATE ION, D-MYO-INOSITOL-4,5-BISPHOSPHATE, GLYCEROL, ... | | Authors: | Egea-Jimenez, A.L, Gallardo, R, Garcia-Pino, A, Ivarsson, Y, Wawrzyniak, A.M, Kashyap, R, Loris, R, Schymkowitz, J, Rousseau, F, Zimmermann, P. | | Deposit date: | 2015-03-30 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the syntenin PDZ1 and PDZ2 tandem in complex with the Frizzled 7 C-terminal fragment and PIP2
To Be Published
|
|
4R7N
 
 | | Fab C2E3 | | Descriptor: | Fab C2E3 Heavy chain, Fab C2E3 Light chain | | Authors: | Loyau, J, Didelot, G, Malinge, P, Ravn, U, Magistrelli, G, Depoisier, J.F, Kosco-Vilbois, M, Fischer, N, Thore, S, Rousseau, F. | | Deposit date: | 2014-08-28 | | Release date: | 2015-07-29 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Robust Antibody-Antigen Complexes Prediction Generated by Combining Sequence Analyses, Mutagenesis, In Vitro Evolution, X-ray Crystallography and In Silico Docking.
J.Mol.Biol., 427, 2015
|
|
4R7D
 
 | | Fab Hu 15C1 | | Descriptor: | Fab Hu 15C1 Heavy chain, Fab Hu 15C1 Light chain | | Authors: | Loyau, J, Didelot, G, Malinge, P, Ravn, U, Magistrelli, G, Depoisier, J.F, Kosco-Vilbois, M, Fischer, N, Thore, S, Rousseau, F. | | Deposit date: | 2014-08-27 | | Release date: | 2015-07-29 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Robust Antibody-Antigen Complexes Prediction Generated by Combining Sequence Analyses, Mutagenesis, In Vitro Evolution, X-ray Crystallography and In Silico Docking.
J.Mol.Biol., 427, 2015
|
|
5FR3
 
 | | X-ray crystal structure of aggregation-resistant protective antigen of Bacillus anthracis (mutant S559L T576E) | | Descriptor: | CALCIUM ION, GLYCEROL, PROTECTIVE ANTIGEN | | Authors: | Ganesan, A, Siekierska, A, Beerten, J, Brams, M, van Durme, J, De Baets, G, van der Kant, R, Gallardo, R, Ramakers, M, Langenberg, T, Wilkinson, H, De Smet, F, Ulens, C, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural Hot Spots for the Solubility of Globular Proteins
Nat.Commun., 7, 2016
|
|
7NXX
 
 | | Structure of Superoxide Dismutase 1 (SOD1) in complex with nanobody 2 (Nb2). | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION, ... | | Authors: | Gallardo, R, Rousseau, F, Schymkowitz, J, Ulens, C. | | Deposit date: | 2021-03-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Identification and rational improvement of a nanobody that suppresses aggregation of mutant SOD1
To Be Published
|
|
6SL6
 
 | | p53 charged core | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION | | Authors: | Gallardo, R, Langenberg, T, Schymkowitz, J, Rousseau, F, Ulens, C. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Thermodynamic and Evolutionary Coupling between the Native and Amyloid State of Globular Proteins.
Cell Rep, 31, 2020
|
|
8OH2
 
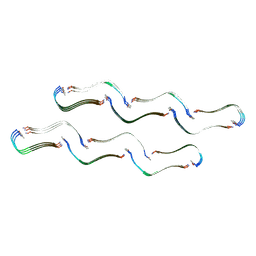 | | Structure of the Tau-PAM4 Type 1 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OHP
 
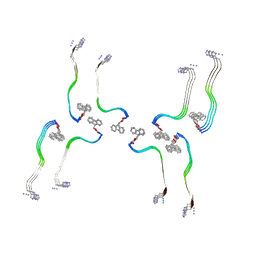 | | Structure of the Fmoc-Tau-PAM4 Type 3 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OI0
 
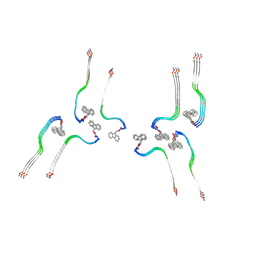 | | Structure of the Fmoc-Tau-PAM4 Type 4 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OHI
 
 | | Structure of the Fmoc-Tau-PAM4 Type 2 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
