1FD6
 
 | |
1FCL
 
 | |
1G28
 
 | |
4TVB
 
 | |
4XR4
 
 | |
4XQE
 
 | |
4XQG
 
 | |
4XQC
 
 | |
4XRG
 
 | |
4XQ9
 
 | |
4PLP
 
 | |
1JNU
 
 | |
3KXE
 
 | |
6ZQT
 
 | | Crystal structure of the RLIP76 Ral binding domain mutant (E427H/Q433L/K440R) in complex with RalB-GMPPNP | | 分子名称: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Hurd, C, Brear, P, Revell, J, Ross, S, Mott, H, Owen, D. | | 登録日 | 2020-07-10 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Affinity maturation of the RLIP76 Ral binding domain to inform the design of stapled peptides targeting the Ral GTPases.
J.Biol.Chem., 296, 2020
|
|
2R13
 
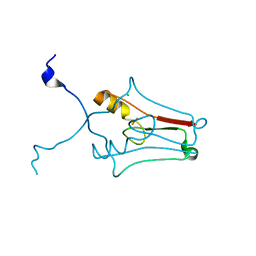 | | Crystal structure of human mitoNEET reveals a novel [2Fe-2S] cluster coordination | | 分子名称: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH domain-containing protein 1 | | 著者 | Hou, X, Liu, R, Ross, S, Smart, E.J, Zhu, H, Gong, W. | | 登録日 | 2007-08-22 | | 公開日 | 2007-09-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic studies of human MitoNEET
J.Biol.Chem., 282, 2007
|
|
2P6J
 
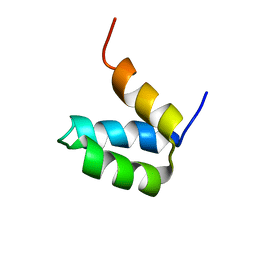 | | Full-sequence computational design and solution structure of a thermostable protein variant | | 分子名称: | designed engrailed homeodomain variant UVF | | 著者 | Shah, P.S, Hom, G.K, Ross, S.A, Lassila, J.K, Crowhurst, K.A, Mayo, S.L. | | 登録日 | 2007-03-18 | | 公開日 | 2007-08-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Full-sequence Computational Design and Solution Structure of a Thermostable Protein Variant.
J.Mol.Biol., 372, 2007
|
|
1LEJ
 
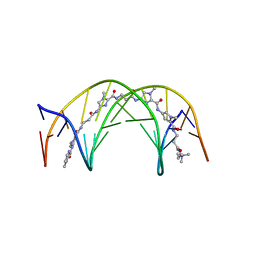 | | NMR Structure of a 1:1 Complex of Polyamide (Im-Py-Beta-Im-Beta-Im-Py-Beta-Dp) with the Tridecamer DNA Duplex 5'-CCAAAGAGAAGCG-3' | | 分子名称: | 5'-D(*CP*CP*AP*AP*AP*GP*AP*GP*AP*AP*GP*CP*G)-3', 5'-D(*CP*GP*CP*TP*TP*CP*TP*CP*TP*TP*TP*GP*G)-3', IMIDAZOLE-PYRROLE-BETA ALANINE-IMIDAZOLE-BETA ALANINE-IMIDAZOLE-PYRROLE-BETA ALANINE-DIMETHYLAMINO PROPYLAMIDE | | 著者 | Urbach, A.R, Love, J.J, Ross, S.A, Dervan, P.B. | | 登録日 | 2002-04-09 | | 公開日 | 2002-05-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a beta-alanine-linked polyamide bound to a full helical turn of purine tract DNA in the 1:1 motif.
J.Mol.Biol., 320, 2002
|
|
6ZRN
 
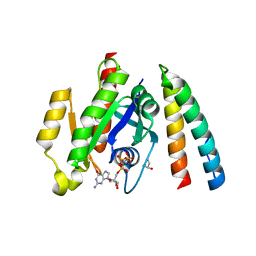 | | Crystal structure of the RLIP76 Ral binding domain mutant (E427S/L429M/Q433L/K440R) in complex with RalB-GMPPNP | | 分子名称: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Hurd, C, Brear, P, Revell, J, Ross, S, Mott, H, Owen, D. | | 登録日 | 2020-07-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.482 Å) | | 主引用文献 | Affinity maturation of the RLIP76 Ral binding domain to inform the design of stapled peptides targeting the Ral GTPases.
J.Biol.Chem., 296, 2020
|
|
1LFM
 
 | | CRYSTAL STRUCTURE OF COBALT(III)-SUBSTITUTED CYTOCHROME C (TUNA) | | 分子名称: | CYTOCHROME C, PROTOPORPHYRIN IX CONTAINING CO | | 著者 | Tezcan, F.A, Findley, W.M, Crane, B.R, Ross, S.A, Lyubovitsky, J.G, Gray, H.B, Winkler, J.R. | | 登録日 | 2002-04-11 | | 公開日 | 2002-07-19 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Using deeply trapped intermediates to map the cytochrome c folding landscape.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4U6S
 
 | | CtBP1 in complex with substrate phenylpyruvate | | 分子名称: | 3-PHENYLPYRUVIC ACID, C-terminal-binding protein 1, CALCIUM ION, ... | | 著者 | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | 登録日 | 2014-07-29 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
4U6Q
 
 | | CtBP1 bound to inhibitor 2-(hydroxyimino)-3-phenylpropanoic acid | | 分子名称: | (2E)-2-(hydroxyimino)-3-phenylpropanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | 著者 | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | 登録日 | 2014-07-29 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
6NTR
 
 | | Crystal Structure of Beta-barrel-like Protein of Domain of Unknown Function DUF1849 from Brucella abortus | | 分子名称: | 1,2-ETHANEDIOL, ATP/GTP-binding site-containing protein A, GLYCEROL | | 著者 | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2019-01-30 | | 公開日 | 2019-02-13 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | BrucellaPeriplasmic Protein EipB Is a Molecular Determinant of Cell Envelope Integrity and Virulence.
J.Bacteriol., 201, 2019
|
|
5K3E
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Glycolate - Cocrystallized | | 分子名称: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID | | 著者 | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | 登録日 | 2016-05-19 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3A
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Both Protomers Reacted with Ligand | | 分子名称: | CHLORIDE ION, Fluoroacetate dehalogenase | | 著者 | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | 登録日 | 2016-05-19 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.511 Å) | | 主引用文献 | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3B
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Chloroacetate - Cocrystallized | | 分子名称: | CHLORIDE ION, Fluoroacetate dehalogenase, chloroacetic acid | | 著者 | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | 登録日 | 2016-05-19 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
