1NL3
 
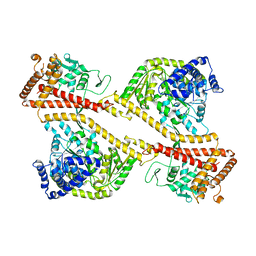 | | CRYSTAL STRUCTURE OF THE SECA PROTEIN TRANSLOCATION ATPASE FROM MYCOBACTERIUM TUBERCULOSIS in APO FORM | | Descriptor: | PREPROTEIN TRANSLOCASE SECA 1 SUBUNIT | | Authors: | Sharma, V, Arockiasamy, A, Ronning, D.R, Savva, C.G, Holzenburg, A, Braunstein, M, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of M. tuberculosis SecA, A Preprotein Translocating ATPase
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1RZ9
 
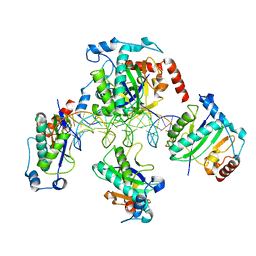 | | Crystal Structure of AAV Rep complexed with the Rep-binding sequence | | Descriptor: | 26-MER, Rep protein | | Authors: | Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M, Dyda, F. | | Deposit date: | 2003-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces.
Mol.Cell, 13, 2004
|
|
4MQL
 
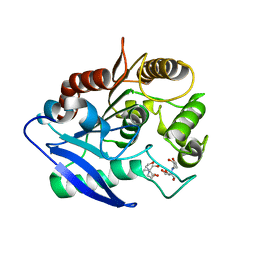 | | Crystal structure of Antigen 85C-C209S mutant | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
1YNK
 
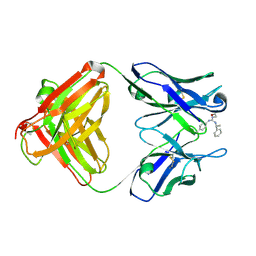 | | Identification of Key residues of the NC6.8 Fab antibody fragment binding to synthetic sweeteners: Crystal structure of NC6.8 co-crystalized with high potency sweetener compound SC45647 | | Descriptor: | 2-[((R)-{[4-(AMINOMETHYL)PHENYL]AMINO}{[(1R)-1-PHENYLETHYL]AMINO}METHYL)AMINO]ETHANE-1,1-DIOL, Ig gamma heavy chain, immunoglobulin kappa light chain | | Authors: | Gokulan, K, Khare, S, Ronning, D.R, Linthicum, S.D, Sacchettini, J.C, Rupp, B. | | Deposit date: | 2005-01-24 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cocrystal Structures of NC6.8 Fab Identify Key Interactions for High Potency Sweetener Recognition: Implications for the Design of Synthetic Sweeteners
Biochemistry, 44, 2005
|
|
1NKT
 
 | | CRYSTAL STRUCTURE OF THE SECA PROTEIN TRANSLOCATION ATPASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEX WITH ADPBS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Preprotein translocase secA 1 subunit | | Authors: | Sharma, V, Arockiasamy, A, Ronning, D.R, Savva, C.G, Holzenburg, A, Braunstein, M, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-01-03 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal Structure of M. tuberculosis SecA, A Preprotein Translocating ATPase
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3HRH
 
 | | Crystal Structure of Antigen 85C and Glycerol | | Descriptor: | Antigen 85-C, GLYCEROL | | Authors: | Boucau, J, Sanki, A.K, Umesiri, F.E, Sucheck, S.J, Ronning, D.R. | | Deposit date: | 2009-06-09 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, synthesis and biological evaluation of sugar-derived esters, alpha-ketoesters and alpha-ketoamides as inhibitors for Mycobacterium tuberculosis antigen 85C.
Mol Biosyst, 5, 2009
|
|
2BZF
 
 | | Structural basis for DNA bridging by barrier-to-autointegration factor (BAF) | | Descriptor: | 5'-D(*CP*CP*TP*CP*CP*AP*CP)-3', 5'-D(*GP*TP*GP*GP*AP*GP*GP)-3', BARRIER-TO-AUTOINTEGRATION FACTOR | | Authors: | Bradley, C.M, Ronning, D.R, Ghirlando, R, Craigie, R, Dyda, F. | | Deposit date: | 2005-08-16 | | Release date: | 2005-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for DNA Bridging by Barrier-to-Autointegration Factor.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1UUT
 
 | | The Nuclease Domain of Adeno-Associated Virus Rep Complexed with the RBE' Stemloop of the Viral Inverted Terminal Repeat | | Descriptor: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*TP*TP*GP *AP*GP*CP*TP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Dyda, F, Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M. | | Deposit date: | 2004-01-10 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Nuclease Domain of Adeno-Associated Virus Rep Coordinates Replication Initiation Using Two Distinct DNA Recognition Interfaces
Mol.Cell, 13, 2004
|
|
1MTO
 
 | | Crystal structure of a Phosphofructokinase mutant from Bacillus stearothermophilus bound with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructokinase | | Authors: | Riley-Lovingshimer, M.R, Ronning, D.R, Sacchettini, J.C, Reinhart, G.D. | | Deposit date: | 2002-09-21 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Reversible Ligand-Induced Dissociation of a Tryptophan-Shift Mutant of Phosphofructokinase
from Bacillus stearothermophilus
Biochemistry, 41, 2002
|
|
2VIH
 
 | | CRYSTAL STRUCTURE OF THE IS608 TRANSPOSASE IN COMPLEX WITH Left END 26-MER DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VHG
 
 | | Crystal Structure of the ISHp608 Transposase in Complex with Right End 31-mer DNA | | Descriptor: | MANGANESE (II) ION, RIGHT END 31-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VJV
 
 | | Crystal structure of the IS608 transposase in complex with left end 26-mer DNA hairpin and a 6-mer DNA representing the left end cleavage site | | Descriptor: | 5'-D(*DA*DA*DA*DG*DC*DC*DC*DC*DT*DA*DG*DC*DTP*DT *DT*DT*DA*DG*DC*DT*DA*DT*DG*DG*DG*DGP)-3', 5'-D(*DT*DA*DT*DT*DA*DCP)-3', MAGNESIUM ION, ... | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VIC
 
 | | CRYSTAL STRUCTURE OF THE ISHP608 TRANSPOSASE IN COMPLEX with Left end 26- mer DNA and manganese | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', MANGANESE (II) ION, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VJU
 
 | | Crystal structure of the IS608 transposase in complex with the complete Right end 35-mer DNA and manganese | | Descriptor: | MANGANESE (II) ION, RIGHT END 35-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
8CNY
 
 | | Structure of Enterovirus A71 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of EV D68 3C protease - to be published
To Be Published
|
|
7HG0
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z46169212 | | Descriptor: | 1,2-ETHANEDIOL, Hepatoma-derived growth factor-related protein 2, N-(4-acetylphenyl)-2-(pyrrolidin-1-yl)acetamide, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
7HG4
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z1696844792 | | Descriptor: | 1,2-ETHANEDIOL, 1-(diphenylmethyl)azetidin-3-ol, Hepatoma-derived growth factor-related protein 2, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
7HG2
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z1401276297 | | Descriptor: | (5S)-7-(pyrazin-2-yl)-2-oxa-7-azaspiro[4.4]nonane, (5~{R})-7-pyrazin-2-yl-2-oxa-7-azaspiro[4.4]nonane, 1,2-ETHANEDIOL, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
7HG3
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z319891284 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2S)-1-(methanesulfonyl)pyrrolidin-2-yl]-5-methyl-1,2-oxazole, Hepatoma-derived growth factor-related protein 2, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
7HGA
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z1980894300 | | Descriptor: | 1,2-ETHANEDIOL, Hepatoma-derived growth factor-related protein 2, SULFATE ION, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
7HGB
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z33297786 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-[(4-methylphenyl)methyl]acetamide, Hepatoma-derived growth factor-related protein 2, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
7HG8
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z445856640 | | Descriptor: | 1,2-ETHANEDIOL, Hepatoma-derived growth factor-related protein 2, N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]-N-methyl-N'-propan-2-ylurea, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
7HG9
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z131516158 | | Descriptor: | 1,2-ETHANEDIOL, Hepatoma-derived growth factor-related protein 2, N-{(1R)-1-[(4S)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}acetamide, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
7HG6
 
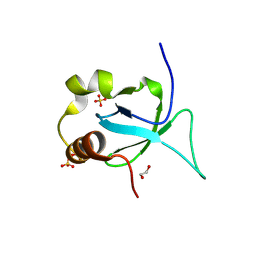 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z839706072 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-bromanylpyrazol-1-yl)-~{N}-cyclopropyl-~{N}-methyl-ethanamide, Hepatoma-derived growth factor-related protein 2, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
7HGD
 
 | | PanDDA analysis group deposition -- Crystal structure of HRP-2 PWWP domain in complex with Z363071686 | | Descriptor: | 1,2-ETHANEDIOL, Hepatoma-derived growth factor-related protein 2, N-methyl-N-[2-oxo-2-(piperidin-1-yl)ethyl]methanesulfonamide, ... | | Authors: | Vantieghem, T, Osipov, E, Fearon, D, Douangamath, A, von Delft, F, Strelkov, S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rational fragment-based design of compounds targeting the PWWP domain of the HRP family.
Eur.J.Med.Chem., 280, 2024
|
|
