2EEM
 
 | |
1CRL
 
 | |
1CIY
 
 | |
1LPS
 
 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1S)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-05 | | Release date: | 1995-02-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPP
 
 | |
1LPO
 
 | |
1LPN
 
 | |
1LPM
 
 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1R)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-06 | | Release date: | 1995-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1TRH
 
 | |
3RAR
 
 | |
2LKQ
 
 | | NMR structure of the lambda 5 22-45 peptide | | Descriptor: | Immunoglobulin lambda-like polypeptide 1 | | Authors: | Elantak, L, Espeli, M, Boned, A, Bornet, O, Breton, C, Feracci, M, Roche, P, Guerlesquin, F, Schiff, C. | | Deposit date: | 2011-10-19 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Galectin-1-dependent Pre-B Cell Receptor (Pre-BCR) Activation.
J.Biol.Chem., 287, 2012
|
|
2BVO
 
 | | Structures of Three HIV-1 HLA-B5703-Peptide Complexes and Identification of Related HLAs Potentially Associated with Long-Term Non-Progression | | Descriptor: | BETA-2-MICROGLOBULIN, GAG PROTEIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B, Gillespie, G, Overton, I.M, Kaul, R, Roche, P, Mcmichael, A.J, Rowland-Jones, S, Jones, E.Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-13 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression.
J Immunol., 175, 2005
|
|
2BVP
 
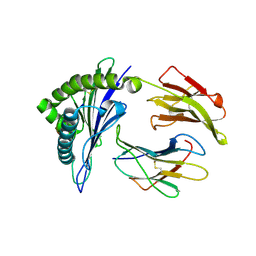 | | Structures of Three HIV-1 HLA-B5703-Peptide Complexes and Identification of Related HLAs Potentially Associated with Long-Term Non-Progression | | Descriptor: | BETA-2-MICROGLOBULIN, HIV-P24, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B, Gillespie, G, Overton, I.M, Kaul, R, Roche, P, Mcmichael, A.J, Rowland-Jones, S, Jones, E.Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-07 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression.
J Immunol., 175, 2005
|
|
2BVQ
 
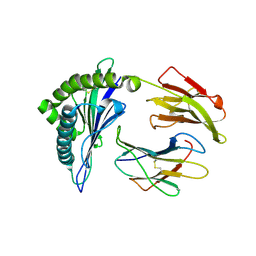 | | Structures of Three HIV-1 HLA-B5703-Peptide Complexes and Identification of Related HLAs Potentially Associated with Long-Term Non-Progression | | Descriptor: | BETA-2-MICROGLOBULIN, HIV-P24, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B, Gillespie, G, Overton, I.M, Kaul, R, Roche, P, Mcmichael, A.J, Rowland-Jones, S, Jones, E.Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-07 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression.
J Immunol., 175, 2005
|
|
1MIR
 
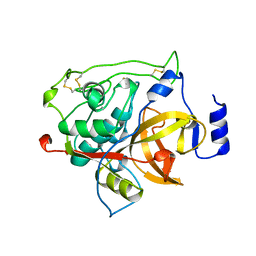 | | RAT PROCATHEPSIN B | | Descriptor: | PROCATHEPSIN B | | Authors: | Cygler, M, Sivaraman, J, Grochulski, P, Coulombe, R, Storer, A.C, Mort, J.S. | | Deposit date: | 1996-01-12 | | Release date: | 1997-01-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion.
Structure, 4, 1996
|
|
7N02
 
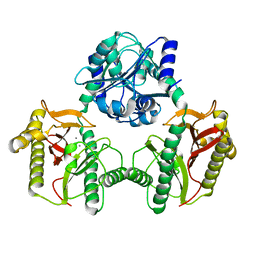 | |
7K3U
 
 | |
4XBH
 
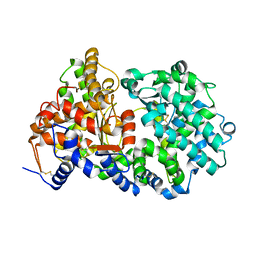 | | Soluble rabbit neprilysin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Labiuk, S.L, Grochulski, P, Sygusch, J. | | Deposit date: | 2014-12-16 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Structures of soluble rabbit neprilysin complexed with phosphoramidon or thiorphan.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6XMR
 
 | |
3H5X
 
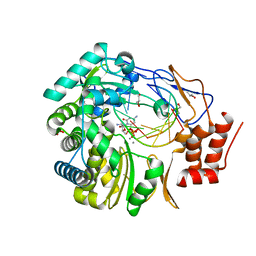 | | Crystal Structure of 2'-amino-2'-deoxy-cytidine-5'-triphosphate bound to Norovirus GII RNA polymerase | | Descriptor: | 2'-amino-2'-deoxycytidine 5'-(tetrahydrogen triphosphate), 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
3H5Y
 
 | | Norovirus polymerase+primer/template+CTP complex at 6 mM MnCl2 | | Descriptor: | 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
4ZR5
 
 | | Soluble rabbit neprilysin in complex with phosphoramidon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Labiuk, S.L, Grochulski, P, Sygusch, J. | | Deposit date: | 2015-05-11 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8016 Å) | | Cite: | Structures of soluble rabbit neprilysin complexed with phosphoramidon or thiorphan.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1AJ5
 
 | | CALPAIN DOMAIN VI APO | | Descriptor: | CALPAIN | | Authors: | Cygler, M, Grochulski, P, Blanchard, H. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
4QPX
 
 | | NV polymerase post-incorporation-like complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Polyprotein, ... | | Authors: | Zamyatkin, D.F, Parra, F, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2014-06-25 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of a backtracked state reveals conformational changes similar to the state following nucleotide incorporation in human norovirus polymerase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1NP8
 
 | | 18-k C-terminally trunucated small subunit of calpain | | Descriptor: | CADMIUM ION, Calcium-dependent protease, small subunit | | Authors: | Leinala, E.K, Arthur, J.S, Grochulski, P, Davies, P.L, Elce, J.S, Jia, Z. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A second binding site revealed by C-terminal truncation of calpain small subunit, a penta-EF-hand protein
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
