7TTR
 
 | | Skd3_ATPyS_FITC-casein Hexamer, AAA+ only | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-casein, Caseinolytic peptidase B protein homolog, ... | | Authors: | Rizo, A.N. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-28 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Unique structural features govern the activity of a human mitochondrial AAA+ disaggregase, Skd3.
Cell Rep, 40, 2022
|
|
7TTS
 
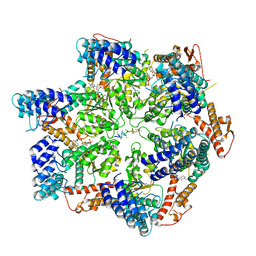 | | Skd3, hexamer, filtered | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-casein, Caseinolytic peptidase B protein homolog, ... | | Authors: | Rizo, A.N, Cupo, R.R. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Unique structural features govern the activity of a human mitochondrial AAA+ disaggregase, Skd3.
Cell Rep, 40, 2022
|
|
8EA3
 
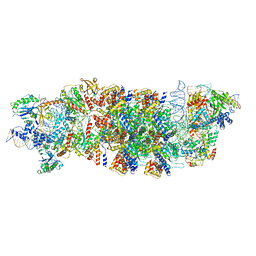 | | V-K CAST Transpososome from Scytonema hofmanni, major configuration | | Descriptor: | 30S ribosomal protein S15, ADENOSINE-5'-TRIPHOSPHATE, Cas12k, ... | | Authors: | Rizo, A.N, Park, J.-U, Tsai, A.W, Kellogg, E.H. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of the holo CRISPR RNA-guided transposon integration complex.
Nature, 613, 2023
|
|
5VY9
 
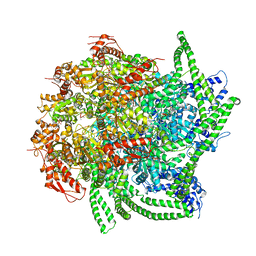 | | S. cerevisiae Hsp104:casein complex, Middle Domain Conformation | | Descriptor: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VY8
 
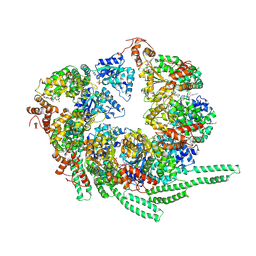 | | S. cerevisiae Hsp104-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 104 | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
5VYA
 
 | | S. cerevisiae Hsp104:casein complex, Extended Conformation | | Descriptor: | Alpha-S1-casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
6UQO
 
 | | ClpA/ClpP Engaged State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp endopeptidase proteolytic subunit ClpP, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Southworth, D.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-04-22 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W20
 
 | | ClpAP Disengaged State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W22
 
 | | ClpA Engaged1 State bound to RepA-GFP (ClpA Focused Refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W1Z
 
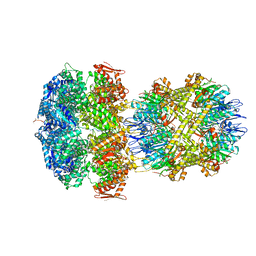 | | ClpAP Engaged1 State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W23
 
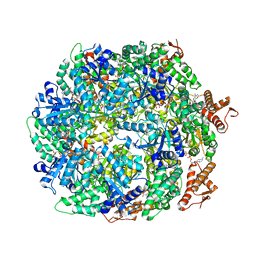 | | ClpA Disengaged State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W21
 
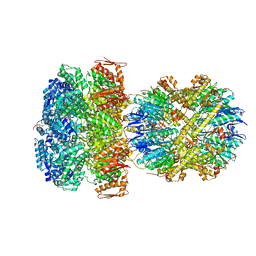 | | ClpAP Engaged2 State bound to RepA-GFP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W24
 
 | | ClpA Engaged2 State bound to RepA-GFP (Focused Classification) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ... | | Authors: | Lopez, K.L, Rizo, A.N, Tse, E, Lin, J, Scull, N.W, Thwin, A.C, Lucius, A.L, Shorter, J, Southworth, D.R. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5VJH
 
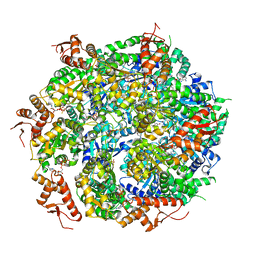 | | Closed State CryoEM Reconstruction of Hsp104:ATPyS and FITC casein | | Descriptor: | FITC casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
