1QJA
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 2) | | Descriptor: | 14-3-3 PROTEIN ZETA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
1AM4
 
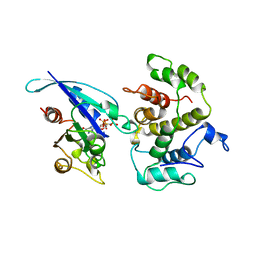 | | COMPLEX BETWEEN CDC42HS.GMPPNP AND P50 RHOGAP (H. SAPIENS) | | Descriptor: | CDC42HS, MAGNESIUM ION, P50-RHOGAP, ... | | Authors: | Rittinger, K, Walker, P, Gamblin, S.J, Smerdon, S.J. | | Deposit date: | 1997-06-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP.
Nature, 388, 1997
|
|
1TX4
 
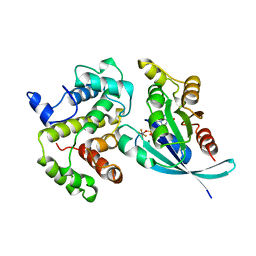 | | RHO/RHOGAP/GDP(DOT)ALF4 COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, P50-RHOGAP, ... | | Authors: | Rittinger, K, Walker, P.A, Smerdon, S.J, Gamblin, S.J. | | Deposit date: | 1997-07-29 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue.
Nature, 389, 1997
|
|
5FEY
 
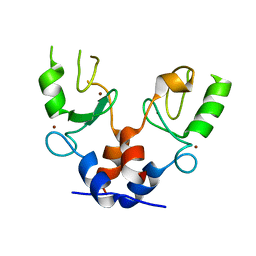 | | TRIM32 RING | | Descriptor: | E3 ubiquitin-protein ligase TRIM32, ZINC ION | | Authors: | Rittinger, K, Esposito, D, Koliopoulos, M.G. | | Deposit date: | 2015-12-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity.
Embo J., 35, 2016
|
|
5FER
 
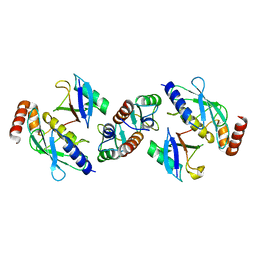 | | Complex of TRIM25 RING with UbcH5-Ub | | Descriptor: | E3 ubiquitin/ISG15 ligase TRIM25, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Rittinger, K, Koliopoulos, M.G, Esposito, D. | | Deposit date: | 2015-12-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity.
Embo J., 35, 2016
|
|
1QJB
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 1) | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
2AK5
 
 | | beta PIX-SH3 complexed with a Cbl-b peptide | | Descriptor: | 8-residue peptide from a signal transduction protein CBL-B, Rho guanine nucleotide exchange factor 7 | | Authors: | Jozic, D, Cardenes, N, Deribe, Y.L, Moncalian, G, Hoeller, D, Groemping, Y, Dikic, I, Rittinger, K, Bravo, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cbl promotes clustering of endocytic adaptor proteins.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7ZJ3
 
 | | Structure of TRIM2 RING domain in complex with UBE2D1~Ub conjugate | | Descriptor: | Polyubiquitin-C, Tripartite motif-containing protein 2, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Esposito, D, Garza-Garcia, A, Dudley-Fraser, J, Rittinger, K. | | Deposit date: | 2022-04-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Divergent self-association properties of paralogous proteins TRIM2 and TRIM3 regulate their E3 ligase activity.
Nat Commun, 13, 2022
|
|
1E96
 
 | | Structure of the Rac/p67phox complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NEUTROPHIL CYTOSOL FACTOR 2 (NCF-2) TPR DOMAIN, ... | | Authors: | Lapouge, K, Smith, S.J.M, Walker, P.A, Gamblin, S.J, Smerdon, S.J, Rittinger, K. | | Deposit date: | 2000-10-10 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the TPR domain of p67phox in complex with Rac.GTP.
Mol.Cell, 6, 2000
|
|
8BVL
 
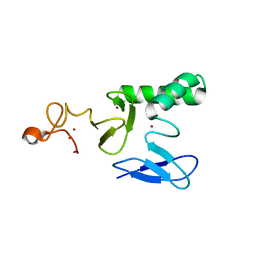 | |
4EMO
 
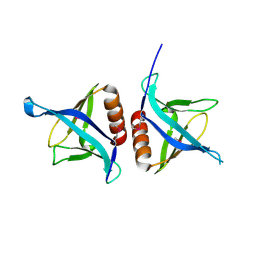 | | Crystal structure of the PH domain of SHARPIN | | Descriptor: | Sharpin | | Authors: | Stieglitz, B, Haire, L.F, Dikic, I, Rittinger, K. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of SHARPIN, a Subunit of a Large Multi-protein E3 Ubiquitin Ligase, Reveals a Novel Dimerization Function for the Pleckstrin Homology Superfold.
J.Biol.Chem., 287, 2012
|
|
6I9H
 
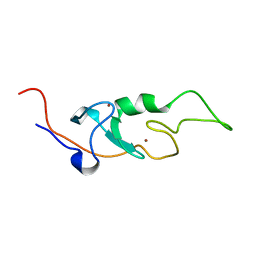 | |
5NT1
 
 | |
5NT2
 
 | |
1OW3
 
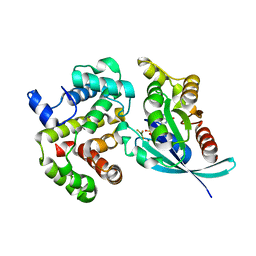 | | Crystal Structure of RhoA.GDP.MgF3-in Complex with RhoGAP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rho-GTPase-activating protein 1, ... | | Authors: | Graham, D.L, Lowe, P.N, Grime, G.W, Marsh, M, Rittinger, K, Smerdon, S.J, Gamblin, S.J, Eccleston, J.F. | | Deposit date: | 2003-03-28 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | MgF(3)(-) as a Transition State Analog of Phosphoryl Transfer
Chem.Biol., 9, 2002
|
|
1OV3
 
 | | Structure of the p22phox-p47phox complex | | Descriptor: | Flavocytochrome b558 alpha polypeptide, Neutrophil cytosol factor 1 | | Authors: | Groemping, Y, Lapouge, K, Smerdon, S.J, Rittinger, K. | | Deposit date: | 2003-03-25 | | Release date: | 2003-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of phosphorylation-induced activation of the NADPH oxidase
Cell(Cambridge,Mass.), 113, 2003
|
|
1NG2
 
 | | Structure of autoinhibited p47phox | | Descriptor: | Neutrophil cytosolic factor 1 | | Authors: | Groemping, Y, Lapouge, K, Smerdon, S.J, Rittinger, K. | | Deposit date: | 2002-12-16 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of phosphorylation-induced activation of the NADPH oxidase
Cell(Cambridge,Mass.), 113, 2003
|
|
4LJO
 
 | | Structure of an active ligase (HOIP)/ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, IMIDAZOLE, Polyubiquitin-C, ... | | Authors: | Rana, R.R, Stieglitz, B, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
2W6B
 
 | |
2W6A
 
 | |
2VRW
 
 | |
4LJP
 
 | | Structure of an active ligase (HOIP-H889A)/ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Polyubiquitin-C, ZINC ION | | Authors: | Rana, R.R, Stieglitz, B, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
4LJQ
 
 | | Crystal structure of the catalytic core of E3 ligase HOIP | | Descriptor: | E3 ubiquitin-protein ligase RNF31, ZINC ION | | Authors: | Stieglitz, B, Rana, R.R, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
8PX1
 
 | | Structure of salmonella effector SseK3, solved at wavelength 2.75 A | | Descriptor: | Non-LEE encoded effector protein NleB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C.M, Esposito, D, Rittinger, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
6EYR
 
 | |
