8DB3
 
 | | Crystal structure of KaiC with truncated C-terminal coiled-coil domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2022-06-14 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8DBA
 
 | | Crystal structure of dodecameric KaiC | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC, MAGNESIUM ION | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2022-06-14 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OU9
 
 | | Asymmetric focused reconstruction of human norovirus GI.7 Houston strain VLP asymmetric unit in T=3 symmetry | | 分子名称: | Major capsid protein | | 著者 | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | 登録日 | 2019-05-04 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OUT
 
 | | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | | 分子名称: | Capsid protein VP1 | | 著者 | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | 登録日 | 2019-05-05 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OUC
 
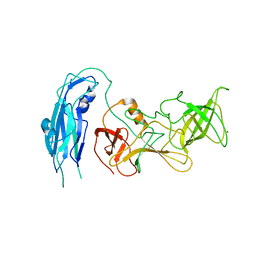 | | Asymmetric focsued reconstruction of human norovirus GII.2 Snow Mountain Virus strain VLP asymmetric unit in T=1 symmetry | | 分子名称: | Viral protein 1, ZINC ION | | 著者 | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | 登録日 | 2019-05-04 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OUU
 
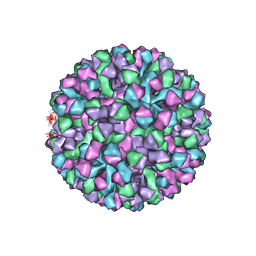 | | Symmetric reconstruction of human norovirus GII.4 Minerva strain VLP in T=4 symmetry | | 分子名称: | Major capsid protein | | 著者 | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | 登録日 | 2019-05-05 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OTF
 
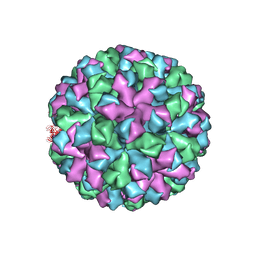 | | Symmetric reconstruction of human norovirus GII.2 Snow Mountain Virus Strain VLP in T=3 symmetry | | 分子名称: | Viral protein 1, ZINC ION | | 著者 | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | 登録日 | 2019-05-03 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8FWI
 
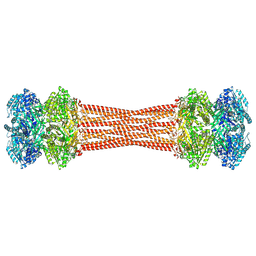 | | Structure of dodecameric KaiC-RS-S413E/S414E solved by cryo-EM | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiC, ... | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2023-01-22 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8FWJ
 
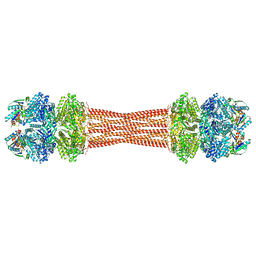 | | Structure of dodecameric KaiC-RS-S413E/S414E complexed with KaiB-RS solved by cryo-EM | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiB, ... | | 著者 | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | 登録日 | 2023-01-22 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
6VWN
 
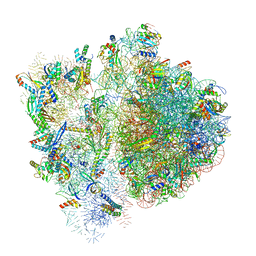 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P-site tRNA (non-rotated conformation, Structure II) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | 登録日 | 2020-02-20 | | 公開日 | 2020-06-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
6AWB
 
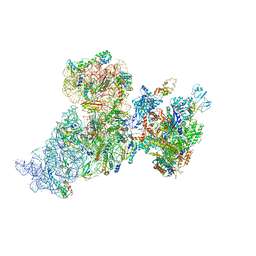 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | 分子名称: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | 登録日 | 2017-09-05 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWD
 
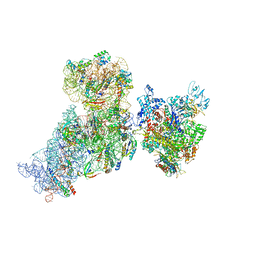 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | 登録日 | 2017-09-05 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (8.1 Å) | | 主引用文献 | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWC
 
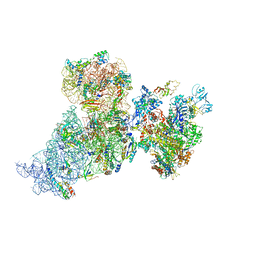 | | Structure of 30S ribosomal subunit and RNA polymerase complex in rotated state | | 分子名称: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | 登録日 | 2017-09-05 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (7.9 Å) | | 主引用文献 | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
5FXJ
 
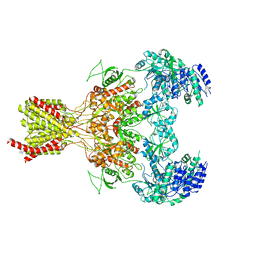 | | GluN1b-GluN2B NMDA receptor structure-Class X | | 分子名称: | GLUTAMATE RECEPTOR IONOTROPIC, NMDA 1, NMDA 2B | | 著者 | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa H, H. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.25 Å) | | 主引用文献 | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXK
 
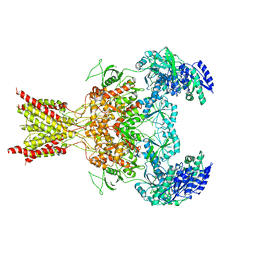 | | GluN1b-GluN2B NMDA receptor structure-Class Y | | 分子名称: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | 著者 | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5J0N
 
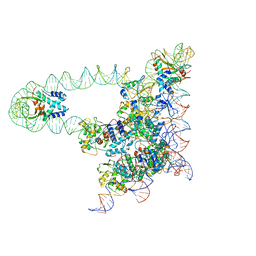 | | Lambda excision HJ intermediate | | 分子名称: | Excisionase, Integrase, Integration host factor subunit alpha, ... | | 著者 | Van Duyne, G, Grigorieff, N, Landy, A. | | 登録日 | 2016-03-28 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Structure of a Holliday junction complex reveals mechanisms governing a highly regulated DNA transaction.
Elife, 5, 2016
|
|
5FXH
 
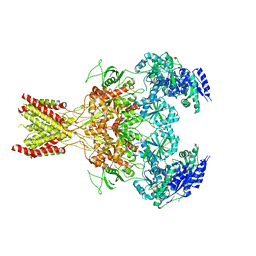 | | GluN1b-GluN2B NMDA receptor in non-active-1 conformation | | 分子名称: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | 著者 | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXG
 
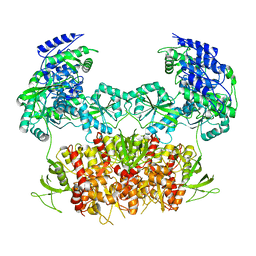 | | GLUN1B-GLUN2B NMDA RECEPTOR IN ACTIVE CONFORMATION | | 分子名称: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | 著者 | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXI
 
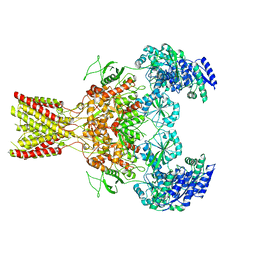 | | GluN1b-GluN2B NMDA receptor structure in non-active-2 conformation | | 分子名称: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | 著者 | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FIL
 
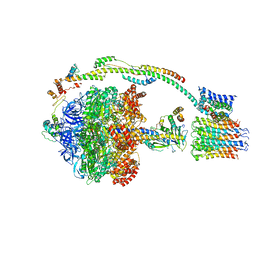 | | Bovine mitochondrial ATP synthase state 3b | | 分子名称: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | 著者 | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | 登録日 | 2015-09-28 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
3J9S
 
 | |
