6BTC
 
 | | SCCmec type IV LP1413 - nucleic acids binding protein | | Descriptor: | LP1413 - SCCmec type IV-encoded DNA binding protein | | Authors: | Rice, P.A, Mir-Sanchis, I. | | Deposit date: | 2017-12-06 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1774075 Å) | | Cite: | Crystal Structure of an Unusual Single-Stranded DNA-Binding Protein Encoded by Staphylococcal Cassette Chromosome Elements.
Structure, 26, 2018
|
|
2NP2
 
 | | Hbb-DNA complex | | Descriptor: | 36-MER, Hbb | | Authors: | Rice, P.A, Mouw, K.W. | | Deposit date: | 2006-10-26 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb.
Mol.Microbiol., 63, 2007
|
|
2RSL
 
 | |
5DGK
 
 | | SCCmec type IV Cch - active helicase | | Descriptor: | Active helicase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Rice, P.A, Mir-Sanchis, I. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Staphylococcal SCCmec elements encode an active MCM-like helicase and thus may be replicative.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1IHF
 
 | | INTEGRATION HOST FACTOR/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (35-MER), DNA (5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*C P*AP*CP*C)-3'), ... | | Authors: | Rice, P.A, Yang, S.-W, Mizuuchi, K, Nash, H.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn.
Cell(Cambridge,Mass.), 87, 1996
|
|
2R0Q
 
 | |
1GDR
 
 | |
1BCM
 
 | |
1BCO
 
 | | BACTERIOPHAGE MU TRANSPOSASE CORE DOMAIN | | Descriptor: | BACTERIOPHAGE MU TRANSPOSASE | | Authors: | Rice, P.A, Mizuuchi, K. | | Deposit date: | 1995-05-26 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration.
Cell(Cambridge,Mass.), 82, 1995
|
|
1P4E
 
 | | Flpe W330F mutant-DNA Holliday Junction Complex | | Descriptor: | 33-MER, 5'-D(*TP*AP*AP*GP*TP*TP*CP*CP*TP*AP*TP*TP*C)-3', 5'-D(*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*TP*AP*GP*GP*AP*AP*CP*TP*TP*C)-3', ... | | Authors: | Rice, P.A, Chen, Y. | | Deposit date: | 2003-04-23 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis
J.Biol.Chem., 278, 2003
|
|
8SH5
 
 | | Crystal structure of 3'cap-independent translation enhancers (CITE) from Pea enation mosaic virus RNA 2 (PEMV2) with Fab BL3-6K170A | | Descriptor: | Fab BL3-6K170A heavy chain, Fab BL3-6K170A light chain, RNA (88-MER) | | Authors: | Lewicka, A, Roman, C, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of a cap-independent translation enhancer RNA.
Nucleic Acids Res., 51, 2023
|
|
3HVT
 
 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | Deposit date: | 1994-07-25 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
6NY9
 
 | | Alpha/beta hydrolase domain-containing protein 10 from mouse | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mycophenolic acid acyl-glucuronide esterase, mitochondrial, ... | | Authors: | Cao, Y, Rice, P.A, Dickinson, B.C. | | Deposit date: | 2019-02-11 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5.
Nat.Chem.Biol., 15, 2019
|
|
4R4P
 
 | | Crystal Structure of the VS ribozyme-A756G mutant | | Descriptor: | MAGNESIUM ION, VS ribozyme RNA | | Authors: | Piccirilli, J.A, Suslov, N.B, Dasgupta, S, Huang, H, Lilley, D.M.J, Rice, P.A. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal structure of the Varkud satellite ribozyme.
Nat.Chem.Biol., 11, 2015
|
|
4R4V
 
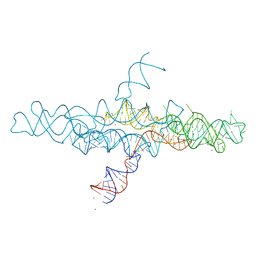 | | Crystal structure of the VS ribozyme - G638A mutant | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, VS ribozyme RNA | | Authors: | Piccirilli, J.A, Suslov, N.B, Dasgupta, S, Huang, H, Lilley, D.M.J, Rice, P.A. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal structure of the Varkud satellite ribozyme.
Nat.Chem.Biol., 11, 2015
|
|
3HSE
 
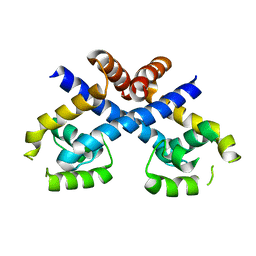 | | Crystal structure of Staphylococcus aureus protein SarZ in reduced form | | Descriptor: | HTH-type transcriptional regulator sarZ | | Authors: | Poor, C.B, Duguid, E, Rice, P.A, He, C. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus.
J.Biol.Chem., 284, 2009
|
|
3HSR
 
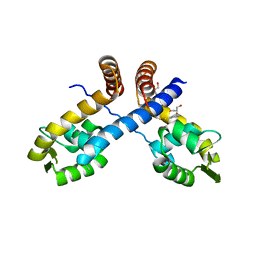 | | Crystal structure of Staphylococcus aureus protein SarZ in mixed disulfide form | | Descriptor: | ACETATE ION, GLYCEROL, HTH-type transcriptional regulator sarZ, ... | | Authors: | Poor, C.B, Duguid, E, Rice, P.A, He, C. | | Deposit date: | 2009-06-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus.
J.Biol.Chem., 284, 2009
|
|
3JSO
 
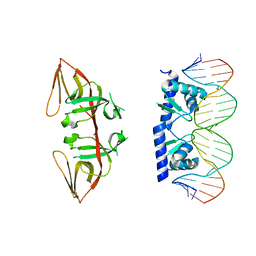 | |
3K3R
 
 | |
3JSP
 
 | |
3HRM
 
 | | Crystal structure of Staphylococcus aureus protein SarZ in sulfenic acid form | | Descriptor: | HTH-type transcriptional regulator sarZ | | Authors: | Poor, C.B, Duguid, E, Rice, P.A, He, C. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus.
J.Biol.Chem., 284, 2009
|
|
3K2Z
 
 | |
8DUZ
 
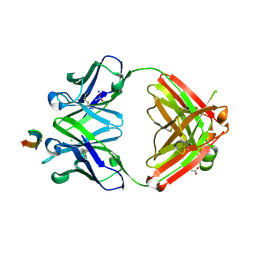 | | Protective antibody against gonococcal lipooligosaccharide bound to peptide mimetic | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Beernink, P.T, Beernink, B.P, Rice, P.A, Ram, S. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protective antibody against gonococcal lipooligosaccharide bound to peptide mimetic
to be published
|
|
4FCY
 
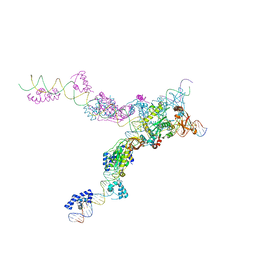 | | Crystal structure of the bacteriophage Mu transpososome | | Descriptor: | DNA (13-MER), DNA (49-MER), DNA (68-MER), ... | | Authors: | Montano, S.P, Pigli, Y.Z, Rice, P.A. | | Deposit date: | 2012-05-25 | | Release date: | 2012-11-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.706 Å) | | Cite: | The Mu transpososome structure sheds light on DDE recombinase evolution.
Nature, 491, 2012
|
|
5C31
 
 | |
