4N4H
 
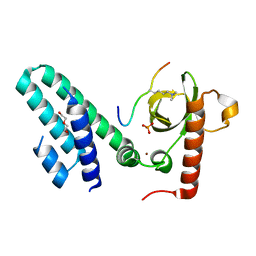 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4N4I
 
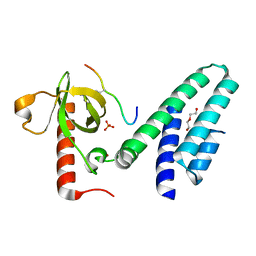 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4N4G
 
 | |
8H3I
 
 | |
8H3J
 
 | |
8GXQ
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in MH-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X, Xu, Y. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8IUE
 
 | | RNA polymerase III pre-initiation complex melting complex 1 | | Descriptor: | DNA (74-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
8IUH
 
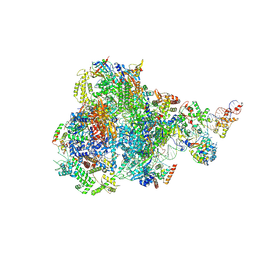 | | RNA polymerase III pre-initiation complex open complex 1 | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
8ITY
 
 | | human RNA polymerase III pre-initiation complex closed DNA 1 | | Descriptor: | DNA (82-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
6ISU
 
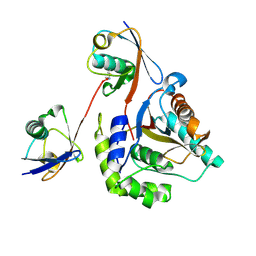 | | Crystal structure of Lys27-linked di-ubiquitin in complex with its selective interacting protein UCHL3 | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | Authors: | Ding, S, Pan, M, Zheng, Q, Ren, Y, Hong, D. | | Deposit date: | 2018-11-19 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | Chemical Protein Synthesis Enabled Mechanistic Studies on the Molecular Recognition of K27-linked Ubiquitin Chains.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
3J7W
 
 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J7X
 
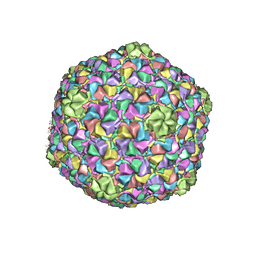 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J7V
 
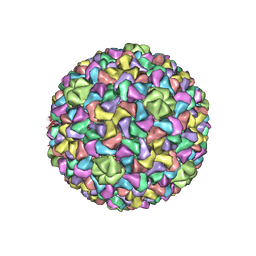 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3MMY
 
 | |
3FHN
 
 | | Structure of Tip20p | | Descriptor: | Protein transport protein TIP20 | | Authors: | Tripathi, A, Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1N4E
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a DNA decamer containing a cis-syn thymine dimer.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SM5
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*(BRU)P*TP*AP*AP*TP*(BRU)P*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2004-03-08 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1T4I
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.S, Kang, C. | | Deposit date: | 2004-04-29 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a cis-syn Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7JV7
 
 | | Crystal Structure of the yeast RNA Pol II CTD kinase CTDK-1 complex | | Descriptor: | CITRATE ANION, CTD kinase subunit alpha, CTD kinase subunit beta, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.850553 Å) | | Cite: | Structure and activation mechanism of the yeast RNA Pol II CTD kinase CTDK-1 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KIR
 
 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant in complex with inositol (1,4)-bisphosphate | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, Inositol polyphosphate 1-phosphatase | | Authors: | Dollins, D.E, Xiong, J.-P, Ren, Y, York, J.D. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
7KIO
 
 | | Crystal structure of inositol polyphosphate 1-phosphatase (INPP1) D54A mutant | | Descriptor: | CALCIUM ION, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Xiong, J.-P, Dollins, D.E, Ren, Y, York, J.D. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
7MD5
 
 | | Insulin receptor ectodomain dimer complexed with two IRPA-9 partial agonists | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
