3M2F
 
 | |
3M23
 
 | |
3M2G
 
 | |
3M2B
 
 | |
3M29
 
 | |
3M2D
 
 | |
8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8XVV
 
 | | The TRRAP module of human NuA4/TIP60 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Isoform 2 of E1A-binding protein p400, Isoform 2 of Transformation/transcription domain-associated protein | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XVG
 
 | | Structure of human NuA4/TIP60 complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, Actin-like protein 6A, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XVT
 
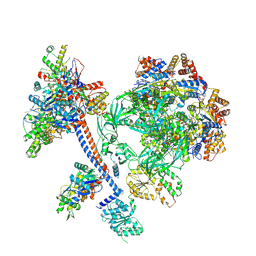 | | The core subcomplex of human NuA4/TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
6E5U
 
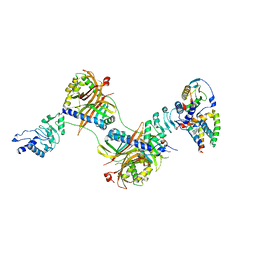 | |
4N4G
 
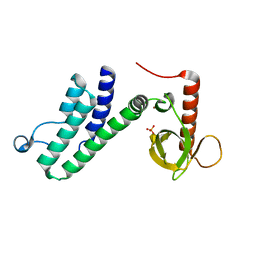 | |
6LNK
 
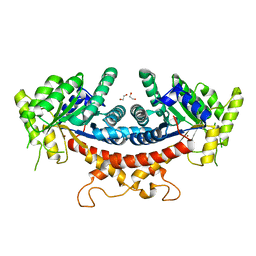 | | Candida albicans Fructose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Huang, Y, Cao, H, Ren, Y, Wan, J. | | Deposit date: | 2019-12-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | Descriptor: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | Descriptor: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2K
 
 | | Crystal structure of nanobody Nb70 with antibody 1F11 fab and SARS-CoV-2 RBD | | Descriptor: | 1F11-H, 1F11-L, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7XI6
 
 | | Crystal structure of C56 from pSSVi | | Descriptor: | C56 | | Authors: | Zhang, Z.F, Ren, Y, Chen, Y.Y, Zhang, X.W, Dong, Y.H, Gong, Y, Cao, P, Huang, L. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the AbrB-like protein C56 conserved in a novel family of integrated genetic elements in Sulfolobales
To Be Published
|
|
8ENK
 
 | |
4TMP
 
 | | Crystal structure of AF9 YEATS bound to H3K9ac peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-LYS-GLN-THR-ALA-ARG-ALY-SER-THR, Protein AF-9 | | Authors: | Li, H, Li, Y, Wang, H, Ren, Y. | | Deposit date: | 2014-06-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Cell, 159, 2014
|
|
6X25
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AND LITHIUM AT 3.2 ANGSTROM RESOLUTION | | Descriptor: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Dollins, D.E, Endo-Streeter, S, Ren, Y, York, J.D. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
7XZG
 
 | | High resolution crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans complexed with NAD+ | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Cao, H, Meng, J, Ren, Y, Wan, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | High resolution crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from Candida albicans complexed with NAD+
To Be Published
|
|
7VS3
 
 | | The crystal structure of rat calcium-dependent activator protein for secretion (CAPS) C2PH | | Descriptor: | Calcium-dependent secretion activator 1, SULFATE ION | | Authors: | Zhou, H, Wei, Z.Q, Zhang, L, Ren, Y.J, Ma, C. | | Deposit date: | 2021-10-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | The C 2 and PH domains of CAPS constitute an effective PI(4,5)P2-binding unit essential for Ca 2+ -regulated exocytosis.
Structure, 31, 2023
|
|
8XN9
 
 | | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6 | | Descriptor: | 1D6 VH, 1D6 VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, P.F, Ren, Y, Yu, C.M, Chen, W. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (1.99 Å) | | Cite: | Nipah virus fusion glycoprotein in complex with a broadly neutralizing antibody 1D6
To Be Published
|
|
