7M29
 
 | |
7M28
 
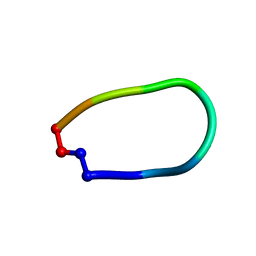 | |
7M27
 
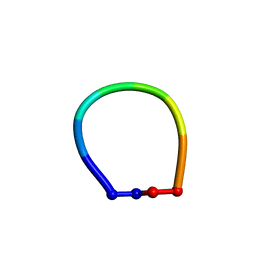 | |
7M2C
 
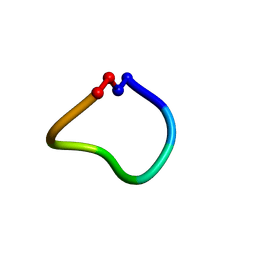 | |
7M2B
 
 | |
7M3U
 
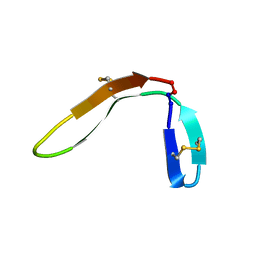 | |
7L53
 
 | |
7L55
 
 | |
2NDN
 
 | | NMR solution structure of PawS Derived Peptide 20 (PDP-20) | | Descriptor: | PawS1a Derived Peptide 20 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-26 | | Release date: | 2016-08-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
2ACQ
 
 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
2MV1
 
 | |
2IXZ
 
 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | Descriptor: | 5'-R(*GP*CP*UP*GP*UP*GP*CP*CP)-3' | | Authors: | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
2IXY
 
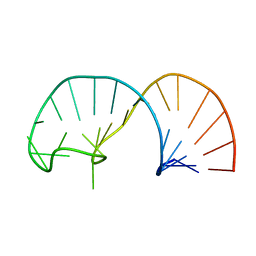 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | Descriptor: | 5'-R(*GP*GP*CP*CP*UP*CP*CP*AP*AP*GP *CP*UP*GP*UP*GP*CP*CP*UP*UP*GP*GP*GP*UP*GP*GP*CP*C)-3' | | Authors: | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
2ACS
 
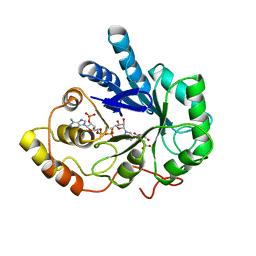 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | ALDOSE REDUCTASE, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
2ACR
 
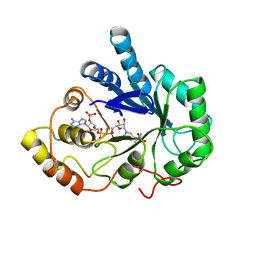 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | ALDOSE REDUCTASE, CACODYLATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
1ZA8
 
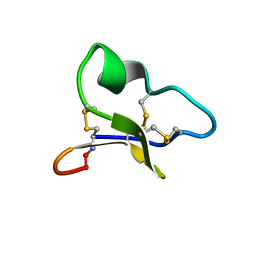 | | NMR solution structure of a leaf-specific-expressed cyclotide vhl-1 | | Descriptor: | vhl-1 | | Authors: | Chen, B, Colgrave, M.L, Daly, N.L, Rosengren, K.J, Gustafson, K.R, Craik, D.J. | | Deposit date: | 2005-04-05 | | Release date: | 2005-04-12 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Isolation and characterization of novel cyclotides from Viola hederaceae: solution structure and anti-HIV activity of vhl-1, a leaf-specific expressed cyclotide.
J.Biol.Chem., 280, 2005
|
|
2MFA
 
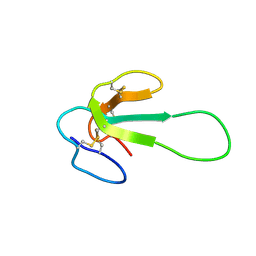 | | Mambalgin-2 | | Descriptor: | Mambalgin-2 | | Authors: | Schroeder, C.I, Rash, L.D, Vila-Farres, X, Rosengren, K.J, Mobli, M, King, G.F, Alewood, P.F, Craik, D.J, Durek, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chemical synthesis, 3D structure, and ASIC binding site of the toxin mambalgin-2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1XGD
 
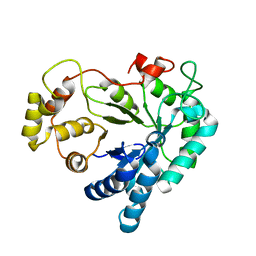 | | Apo R268A human aldose reductase | | Descriptor: | Aldose reductase | | Authors: | Brownlee, J.M, Bohren, K.M, Milne, A.C, Gabbay, K.H, Harrison, D.H.T. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of Apo R268A human aldose reductase: Hinges and latches that control the kinetic mechanism
Biochim.Biophys.Acta, 1748, 2005
|
|
2X6M
 
 | | Structure of a single domain camelid antibody fragment in complex with a C-terminal peptide of alpha-synuclein | | Descriptor: | ALPHA-SYNUCLEIN PEPTIDE, HEAVY CHAIN VARIABLE DOMAIN FROM DROMEDARY | | Authors: | DeGenst, E, Guilliams, T, Wellens, J, O'Day, E.M, Waudby, C.A, Meehan, S, Dumoulin, M, Hsu, S.-T.D, Cremades, N, Verschueren, K.H.G, Pardon, E, Wyns, L, Steyaert, J, Christodoulou, J, Dobson, C.M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Properties of a Complex of Alpha-Synuclein and a Single-Domain Camelid Antibody.
J.Mol.Biol., 402, 2010
|
|
2Q3N
 
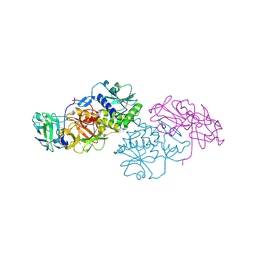 | | Agglutinin from Abrus Precatorius (APA-I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Agglutinin-1 A chain, Agglutinin-1 B chain | | Authors: | Bagaria, A, Surendranath, K, Ramagopal, U.A, Ramakumar, S, Karande, A.A. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Function Analysis and Insights into the Reduced Toxicity of Abrus precatorius Agglutinin I in Relation to Abrin.
J.Biol.Chem., 281, 2006
|
|
2NB6
 
 | |
2MUB
 
 | |
2NB5
 
 | | NMR solution structure of PawS Derived Peptide 9 (PDP-9) | | Descriptor: | Preproalbumin PawS1 | | Authors: | Armstrong, D.A, Franke, B, Elliott, A.G, Mylne, J.S, Rosengren, K.J. | | Deposit date: | 2016-01-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Natural structural diversity within a conserved cyclic peptide scaffold.
Amino Acids, 49, 2017
|
|
1R0Q
 
 | | Characterization of the conversion of the malformed, recombinant cytochrome rc552 to a 2-formyl-4-vinyl (Spirographis) heme | | Descriptor: | 2-FORMYL-PROTOPORPHRYN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
1QYZ
 
 | | Characterization of the malformed, recombinant cytochrome rC552 | | Descriptor: | 2-ACETYL-PROTOPORPHYRIN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
