4TRD
 
 | |
4ZUJ
 
 | |
6OVD
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC | | Descriptor: | (3S,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(3-ethylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
6OVE
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-propylphenyl-ACEPC | | Descriptor: | (3R,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(4-propylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
5C4Z
 
 | |
5C3W
 
 | |
5C4H
 
 | |
7NOZ
 
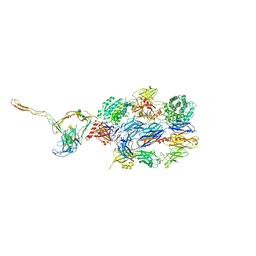 | | Structure of the nanobody stablized properdin bound alternative pathway proconvertase C3b:FB:FP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Lorenzen, J, Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2021-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure determination of an unstable macromolecular complex enabled by nanobody-peptide bridging.
Protein Sci., 31, 2022
|
|
4ZQ3
 
 | |
4ZUI
 
 | |
6FJ3
 
 | | High resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist. | | Descriptor: | ACETIC ACID, CHLORIDE ION, OLEIC ACID, ... | | Authors: | Ehrenmann, J, Schoppe, J, Klenk, C, Rappas, M, Kummer, L, Dore, A.S, Pluckthun, A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4KHV
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23S at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Sorenson, J.L, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2013-05-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23S at cryogenic temperature
TO BE PUBLISHED
|
|
6C07
 
 | |
6ASV
 
 | | E. coli PRPP Synthetase | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Ribose-phosphate pyrophosphokinase | | Authors: | French, J.B, Zhou, W. | | Deposit date: | 2017-08-25 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of E. coli PRPP synthetase.
BMC Struct. Biol., 19, 2019
|
|
4Z35
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4WOC
 
 | | Proteinase-K Post-Surface Acoustic Waves | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WO9
 
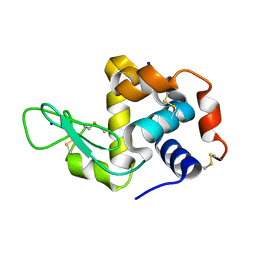 | | Lysozyme Post-Surface Acoustic Waves | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WOB
 
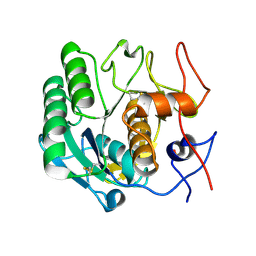 | | Proteinase-K Pre-Surface Acoustic Wave | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WO6
 
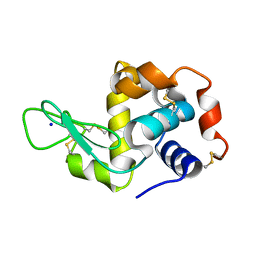 | | Lysozyme Pre-surface acoustic wave | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WOA
 
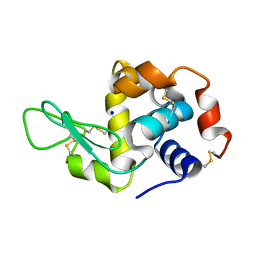 | |
7BF2
 
 | | Ca2+-Calmodulin in complex with human muscle form creatine kinase peptide in extended 1:2 binding mode | | Descriptor: | CALCIUM ION, Calmodulin-1, Creatine kinase M-type | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
7BF1
 
 | | Ca2+-Calmodulin in complex with peptide from brain-type creatine kinase in extended 1:2 binding mode | | Descriptor: | ACETYL GROUP, CALCIUM ION, Calmodulin-1, ... | | Authors: | Sprenger, J, Akerfeldt, K.S, Bredfelt, J, Patel, N, Rowlett, R, Trifan, A, Vanderbeck, A, Lo Leggio, L, Snogerup Linse, S. | | Deposit date: | 2020-12-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Calmodulin complexes with brain and muscle creatine kinase peptides.
Curr Res Struct Biol, 3, 2021
|
|
6HY1
 
 | | Plasmodium falciparum spermidine synthase in complex with 5'-methylthioadenosine and N,N'-Bis(3-aminopropyl)-1,4-cyclohexanediamine after catalysis in crystal | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, ... | | Authors: | Sprenger, J, Coertzen, D, Persson, L, Carey, J, Birkholtz, L.M, Louw, B.I. | | Deposit date: | 2018-10-19 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Plasmodium falciparum spermidine synthase in complex with 5'-methylthioadenosine and N,N'-Bis(3-aminopropyl)-1,4-cyclohexanediamine after catalysis in crystal
To Be Published
|
|
4FF8
 
 | | Inhibitor bound structure of the kinase domain of the murine receptor tyrosine kinase TYRO3 (Sky) | | Descriptor: | 4-(cyclopentylamino)-2-[(2-methoxybenzyl)amino]-N-[3-(2-oxopyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Tyrosine-protein kinase receptor TYRO3 | | Authors: | Ohren, J.F, Powell, N.A, Kohrt, J, Perrin, L.A. | | Deposit date: | 2012-05-31 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly selective 2,4-diaminopyrimidine-5-carboxamide inhibitors of Sky kinase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6I1N
 
 | | Plasmodium falciparum spermidine synthase in complex with N-(3-aminopropyl)-trans-cyclohexane-1,4-diamine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, Spermidine synthase, ... | | Authors: | Sprenger, J, Coertzen, D, Persson, L, Carey, J, Birkholtz, L.M, Louw, B.I. | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85000134 Å) | | Cite: | Plasmodium falciparum spermidine synthase in complex with N-(3-aminopropyl)-trans-cyclohexane-1,4-diamine
To Be Published
|
|
