6WFQ
 
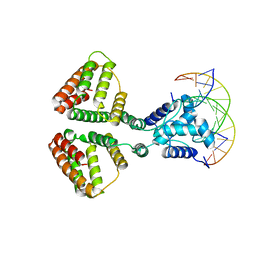 | | NanR dimer-DNA hetero-complex | | Descriptor: | DNA (5'-D(P*GP*GP*TP*AP*TP*AP*AP*CP*AP*GP*GP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*CP*CP*TP*GP*TP*TP*AP*TP*AP*CP*C)-3'), HTH-type transcriptional repressor NanR | | Authors: | Hariprasad, V, Horne, C, Santosh, P, Amy, H, Emre, B, Rachel, N, Michael, G, Georg, R, Borries, D, Renwick, D. | | Deposit date: | 2020-04-03 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism.
Nat Commun, 12, 2021
|
|
7PBU
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvA-HJ core [t2 dataset] | | Descriptor: | Holliday junction, Holliday junction ATP-dependent DNA helicase RuvA | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBT
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s1 [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Wald, J, Fahrenkamp, D, Goessweiner-Mohr, N, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBQ
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBS
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBR
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0-A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, MAGNESIUM ION, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
4HEP
 
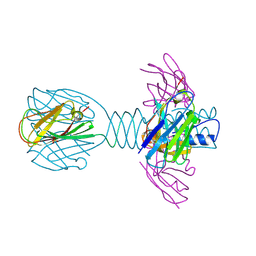 | | Complex of lactococcal phage TP901-1 with a llama vHH (vHH17) binder (nanobody) | | Descriptor: | BPP, SULFATE ION, vHH17 domain | | Authors: | Desmyter, A, Spinelli, S, Farenc, C, Blangy, S, Bebeacua, C, van Sinderen, D, Mahony, J, Cambillau, C. | | Deposit date: | 2012-10-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HEM
 
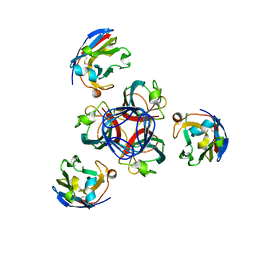 | | Llama vHH-02 binder of ORF49 (RBP) from lactococcal phage TP901-1 | | Descriptor: | Anti-baseplate TP901-1 Llama vHH 02, BPP | | Authors: | Desmyter, A, Spinelli, S, Farenc, C, Blangy, S, Bebeacua, C, van Sinderen, D, Mahony, J, Cambillau, C. | | Deposit date: | 2012-10-04 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8A9O
 
 | | Structure of the polyamine acetyltransferase DpA | | Descriptor: | ACETYL COENZYME *A, BROMIDE ION, COENZYME A, ... | | Authors: | Garcia-Pino, A, Jurenas, D. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.587 Å) | | Cite: | A polyamine acetyltransferase regulates the motility and biofilm formation of Acinetobacter baumannii.
Nat Commun, 14, 2023
|
|
8A9N
 
 | | Structure of DpA polyamine acetyltransferase in complex with 1,3-DAP | | Descriptor: | 1,3-DIAMINOPROPANE, Acetyltransferase, COENZYME A, ... | | Authors: | Garcia-Pino, A, Jurenas, D. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | A polyamine acetyltransferase regulates the motility and biofilm formation of Acinetobacter baumannii.
Nat Commun, 14, 2023
|
|
4IOS
 
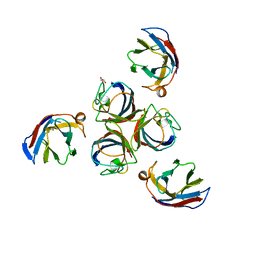 | | Structure of phage TP901-1 RBP (ORF49) in complex with nanobody 11. | | Descriptor: | BPP, GLYCEROL, Llama nanobody 11 | | Authors: | Desmyter, A, Farenc, C, Mahony, J, Spinelli, S, Bebeacua, C, Blangy, S, Veesler, D, van Sinderen, D, Cambillau, C. | | Deposit date: | 2013-01-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7AGX
 
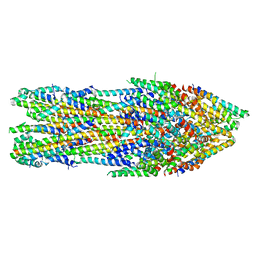 | | Apo-state type 3 secretion system export apparatus complex from Salmonella enterica typhimurium | | Descriptor: | Protein PrgI, Protein PrgJ, Surface presentation of antigens protein SpaP, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation.
Nat Commun, 12, 2021
|
|
6GA3
 
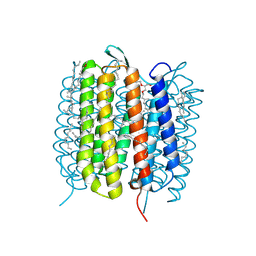 | | Bacteriorhodopsin, 33 ms state, ensemble refinement | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GA1
 
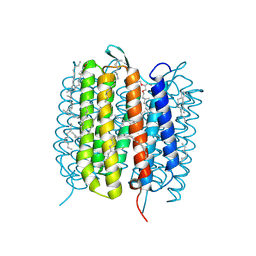 | | Bacteriorhodopsin, dark state, cell 1 | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GA2
 
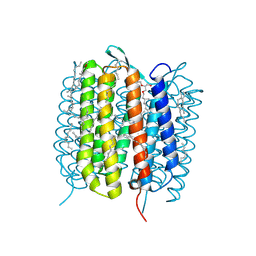 | | Bacteriorhodopsin, dark state, cell 2 | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
7P8K
 
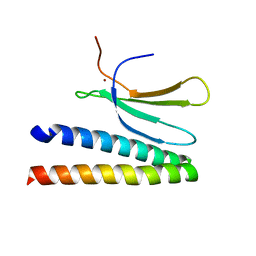 | | Crystal structure of in planta processed AvrRps4 in complex with the WRKY domain of RRS1 | | Descriptor: | Avirulence protein,Avirulence protein, Disease resistance protein RRS1, ZINC ION | | Authors: | Mukhi, N, Brown, H, Gorenkin, D, Ding, P, Bentham, A.R, Jones, J.D.G, Banfield, M.J. | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6QSM
 
 | |
6SC4
 
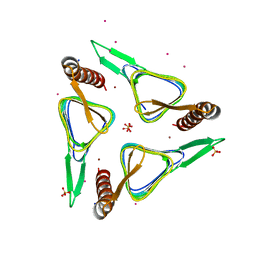 | | Gamma-Carbonic Anhydrase from the Haloarchaeon Halobacterium sp. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Vogler, M, Karan, R, Renn, D, Vancea, A, Vielberg, V.-T, Groetzinger, S.W, DasSarma, P, Das Sarma, S, Eppinger, J, Groll, M, Rueping, M. | | Deposit date: | 2019-07-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Active Site Engineering of a Halophilic gamma-Carbonic Anhydrase.
Front Microbiol, 11, 2020
|
|
8F8W
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, afucosylated IgG1 fragment | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8F8X
 
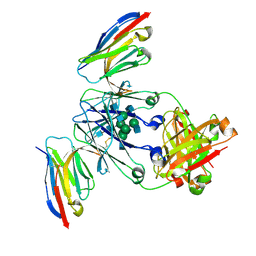 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, Uncharacterized protein DKFZp686C11235 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
4D0W
 
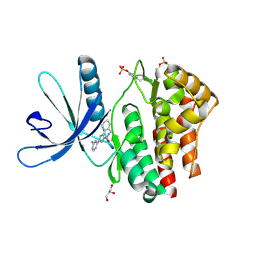 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-(5-chloro-2-methylphenyl)-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4D1S
 
 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 2-(5-chloro-2-methylphenyl)-1-methyl-5-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1H-pyrrole-3-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
4D0X
 
 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-[2-chloro-5-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Canevari, G, Fasolini, M, Bertrand, J, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
3R2J
 
 | | Crystal Structure of PnC1 from L. infantum in complex with nicotinate | | Descriptor: | Alpha/beta-hydrolase-like protein, CHLORIDE ION, NICOTINIC ACID, ... | | Authors: | Gazanion, E, Garcia, D, Guichou, J.-F, Labesse, G, Sereno, D, Vergnes, B. | | Deposit date: | 2011-03-14 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The Leishmania nicotinamidase is essential for NAD(+) production and parasite proliferation.
Mol.Microbiol., 82, 2011
|
|
5E7T
 
 | | Structure of the tripod (BppUct-A-L) from the baseplate of bacteriophage Tuc2009 | | Descriptor: | CALCIUM ION, Major structural protein 1, Minor structural protein 4, ... | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
