5DUN
 
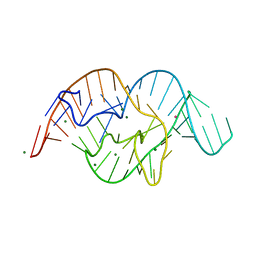 | | The crystal structure of OMe substituted twister ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (54-MER) | | Authors: | Ren, A, Patel, D.J, Micura, R, Rajashankar, K.R. | | Deposit date: | 2015-09-19 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5DDO
 
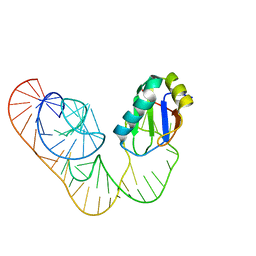 | |
4IX7
 
 | |
3KD7
 
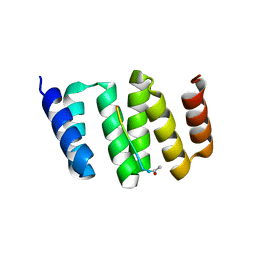 | |
3CB0
 
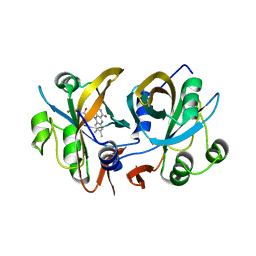 | | CobR | | Descriptor: | 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE, FLAVIN MONONUCLEOTIDE | | Authors: | Lawrence, A.D, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification, characterization, and structure/function analysis of a corrin reductase involved in adenosylcobalamin biosynthesis
J.Biol.Chem., 283, 2008
|
|
3DPA
 
 | |
1I4V
 
 | |
1E50
 
 | | AML1/CBFbeta complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT, CORE-BINDING FACTOR CBF-BETA | | Authors: | Warren, A.J, Bravo, J, Williams, R.L, Rabbits, T.H. | | Deposit date: | 2000-07-13 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Heterodimeric Interaction between the Acute Leukaemia-Associated Transcription Factors Aml1 and Cbfbeta
Embo J., 19, 2000
|
|
4IRA
 
 | | CobR in complex with FAD | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Lawrence, A.D, Scott, A.F, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2013-01-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biophysical characterisation and structure-function analysis of Brucella melitensis CobR: Protein-flavin interactions determine function and stability
To be Published
|
|
1ZEM
 
 | |
1PZ0
 
 | |
1QL3
 
 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the reduced state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-06 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
1QL4
 
 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the oxidised state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
1QLE
 
 | |
1E1D
 
 | | Hybrid Cluster Protein from Desulfovibrio vulgaris | | Descriptor: | HYDROXYLAMINE REDUCTASE, IRON/SULFUR CLUSTER, IRON/SULFUR/OXYGEN HYBRID CLUSTER | | Authors: | Arendsen, A.F, Cooper, S.J, Bailey, S, Garner, C.D, Hagen, W.R, Lindley, P.F. | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Hybrid-Cluster Protein (Hcp) from Desulfovibrio Vulgaris (Hildenborough) at 1.6 A Resolution.
Biochemistry, 39, 2000
|
|
8FY8
 
 | | 5-MeO-DMT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | 2-(5-methoxy-1H-indol-3-yl)-N,N-dimethylethan-1-amine, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-25 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
1PYF
 
 | |
1PZ1
 
 | |
6OEC
 
 | | Yeast Spc42 Trimeric Coiled-Coil Amino Acids 181-211 fused to PDB: 3H5I | | Descriptor: | CALCIUM ION, Response regulator/sensory box protein/GGDEF domain protein,Spindle pole body component SPC42 | | Authors: | Drennan, A.C, Shivaani, K, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jasperson, S.L, Rayment, I. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
6OEI
 
 | | Yeast Spc42 N-terminal coiled-coil fused to PDB: 3K2N | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Spindle pole body component SPC42,Sigma-54-dependent transcriptional regulator | | Authors: | Drennan, A.C, Krishna, S, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jaspersen, S.L, Rayment, I. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
8FYE
 
 | | 4-F, 5-MeO-PyrT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | 4-fluoro-5-methoxy-3-[2-(pyrrolidin-1-yl)ethyl]-1H-indole, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
8FYL
 
 | | Vilazodone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | 5-{4-[4-(5-cyano-1H-indol-3-yl)butyl]piperazin-1-yl}-1-benzofuran-2-carboxamide, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
8FYT
 
 | | LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
8FYX
 
 | | Buspirone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex | | Descriptor: | Buspirone, CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Warren, A.L, Zilberg, G, Capper, M.J, Wacker, D. | | Deposit date: | 2023-01-26 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structural pharmacology and therapeutic potential of 5-methoxytryptamines.
Nature, 630, 2024
|
|
6OD2
 
 | | Yeast Spc42 C-terminal Antiparallel Coiled-Coil | | Descriptor: | Spindle pole body component SPC42 | | Authors: | Drennan, A.C, Krishna, S, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jaspersen, S.L, Rayment, I. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
