3B7Z
 
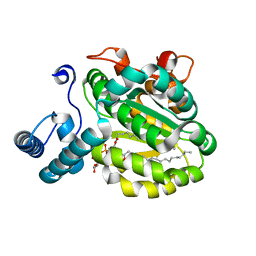 | | Crystal Structure of Yeast Sec14 Homolog Sfh1 in Complex with Phosphatidylcholine or Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Uncharacterized protein YKL091C | | Authors: | Ortlund, E.A, Schaaf, G, Redinbo, M.R, Bankaitis, V. | | Deposit date: | 2007-10-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily
Mol.Cell, 29, 2008
|
|
5I7J
 
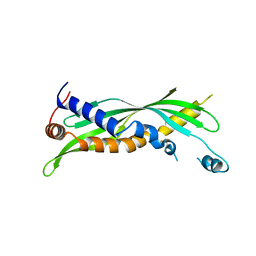 | |
5I7K
 
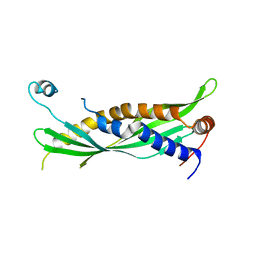 | | Crystal Structure of Human SPLUNC1 Dolphin Mutant D1 (G58A, S61A, G62E, G63D, G66D, I67T) | | Descriptor: | BPI fold-containing family A member 1 | | Authors: | Walton, W.G, Redinbo, M.R. | | Deposit date: | 2016-02-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Structural Features Essential to the Antimicrobial Functions of Human SPLUNC1.
Biochemistry, 55, 2016
|
|
5I7L
 
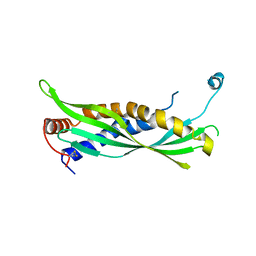 | |
5FF5
 
 | | Crystal Structure of SeMet PaaA | | Descriptor: | GLYCEROL, NICKEL (II) ION, PaaA, ... | | Authors: | Biernat, K.B, Redinbo, M.R. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-27 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (2.933 Å) | | Cite: | Post-translational Claisen Condensation and Decarboxylation en Route to the Bicyclic Core of Pantocin A.
J.Am.Chem.Soc., 138, 2016
|
|
3FLD
 
 | |
5UJ6
 
 | |
4K59
 
 | |
1NH3
 
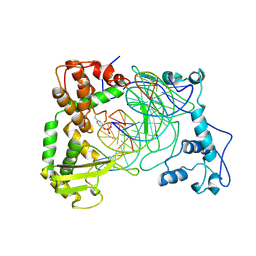 | | Human Topoisomerase I Ara-C Complex | | Descriptor: | 5'-D(*(GNG)P*GP*AP*AP*AP*AP*AP*UP*UP*UP*UP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*UP*(UBB))-3', 5'-D(*AP*AP*AP*AP*AP*TP*UP*UP*UP*UP*CP*(CAR)P*AP*AP*GP*UP*CP*UP*UP*UP*UP*T)-3', ... | | Authors: | Chrencik, J.E, Burgin, A.B, Pommier, Y, Stewart, L, Redinbo, M.R. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Impact of the Leukemia Drug 1-beta-D-Arabinofuranosylcytosine (Ara-C) on the Covalent Human Topoisomerase I-DNA Complex
J.Biol.Chem., 278, 2003
|
|
2DQZ
 
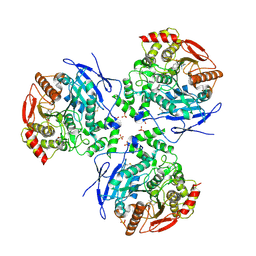 | | Crystal structure of human carboxylesterase in complex with homatropine, coenzyme A, and palmitate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COENZYME A, FLUORIDE ION, ... | | Authors: | Bencharit, S, Redinbo, M.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1
J.Mol.Biol., 363, 2006
|
|
2DQY
 
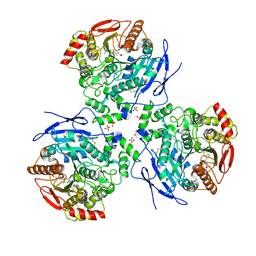 | | Crystal structure of human carboxylesterase in complex with cholate and palmitate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLIC ACID, Liver carboxylesterase 1, ... | | Authors: | Bencharit, S, Edwards, C.C, Morton, C.L, Howard-Williams, E.L, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1
J.Mol.Biol., 363, 2006
|
|
2DR0
 
 | |
6U7I
 
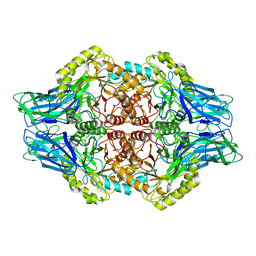 | | Faecalibacterium prausnitzii Beta-glucuronidase | | Descriptor: | Beta-glucuronidase | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gut microbial beta-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens.
J.Biol.Chem., 294, 2019
|
|
6USS
 
 | |
6UST
 
 | | Gut microbial sulfatase from Hungatella hathewayi | | Descriptor: | CALCIUM ION, N-acetylgalactosamine 6-sulfate sulfatase | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Endobiotic Reactivation by Human Gut Microbiome-Encoded Sulfatases.
Biochemistry, 59, 2020
|
|
3R8D
 
 | |
3R0Q
 
 | |
8E72
 
 | | Treponema lecithinolyticum beta-glucuronidase in complex with a ciprofloxacin-glucuronide conjugate | | Descriptor: | 3-carboxy-1-cyclopropyl-6-fluoro-7-(4-beta-D-glucopyranuronosyl-3,4-dihydropyrazin-1(2H)-yl)-4-oxo-1,4-dihydroquinoline, Glycosyl hydrolase family 2, TIM barrel domain protein, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-08-23 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8ESI
 
 | | Bile Salt Hydrolase from B. longum with covalent inhibitor bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Conjugated bile acid hydrolase | | Authors: | Walker, M.E, Lim, L, Redinbo, M.R. | | Deposit date: | 2022-10-14 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ESG
 
 | | Bile Salt Hydrolase B from Lactobacillus gasseri with covalent inhibitor bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Choloylglycine hydrolase | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-14 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ESL
 
 | | Bile Salt Hydrolase from a Bacteroidales species with covalent inhibitor bound | | Descriptor: | (1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-1-[(2R)-6-fluoro-5-oxohexan-2-yl]-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-7-yl hydrogen sulfate (non-preferred name), Choloylglycine hydrolase | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-14 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ETK
 
 | | Bile salt hydrolase A from Lactobacillus gasseri bound to covalent probe | | Descriptor: | (5R)-5-[(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-9a,11a-dimethyl-7-(2-{2-[(prop-2-yn-1-yl)oxy]ethoxy}ethoxy)hexadecahydro-1H-cyclopenta[a]phenanthren-1-yl]-1-fluorohexan-2-one (non-preferred name), Conjugated bile salt hydrolase, SODIUM ION | | Authors: | Walker, M.E, Grundy, M.K, Redinbo, M.R. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ETE
 
 | | Bile Salt Hydrolase from B. longum with covalent inhibitor bound | | Descriptor: | (5R)-1-fluoro-5-[(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-7-hydroxy-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-1-yl]hexan-2-one (non-preferred name), Conjugated bile acid hydrolase | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8ETF
 
 | | Bile Salt Hydrolase B from Lactobacillus gasseri with covalent inhibitor bound | | Descriptor: | (5R)-1-fluoro-5-[(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-7-hydroxy-9a,11a-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-1-yl]hexan-2-one (non-preferred name), Choloylglycine hydrolase, NICKEL (II) ION | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
8EWT
 
 | | Bile salt hydrolase A from Lactobacillus gasseri bound to covalent probe | | Descriptor: | (5R)-5-{(1R,3aS,3bR,5aR,7R,9aS,9bS,11aR)-9a,11a-dimethyl-7-[(prop-2-yn-1-yl)oxy]hexadecahydro-1H-cyclopenta[a]phenanthren-1-yl}-1-fluorohexan-2-one (non-preferred name), Conjugated bile salt hydrolase, SODIUM ION | | Authors: | Walker, M.E, Redinbo, M.R. | | Deposit date: | 2022-10-24 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural diversity of bile salt hydrolases reveals rationale for substrate selectivity
To Be Published
|
|
