7TQV
 
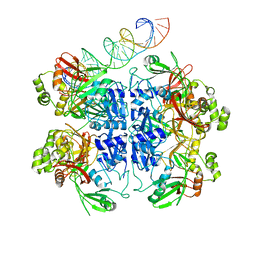 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (33-MER), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
7TJ2
 
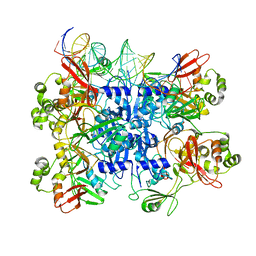 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (31-MER), Uridylate-specific endoribonuclease nsp15 | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
7N06
 
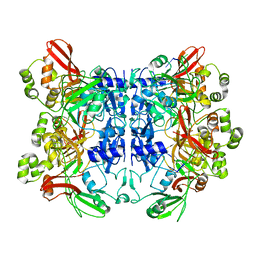 | | SARS-CoV-2 Nsp15 endoribonuclease post-cleavage state | | Descriptor: | RNA (5'-R(*AP*UP*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
7N33
 
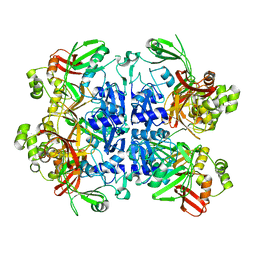 | | SARS-CoV-2 Nsp15 endoribonuclease pre-cleavage state | | Descriptor: | RNA (5'-R(*A)-D(*(UFT))-R(P*A)-3'), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Dillard, L.B, Krahn, J.M, Stanley, R.E. | | Deposit date: | 2021-05-31 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U.
Nucleic Acids Res., 49, 2021
|
|
4DO2
 
 | |
5WF2
 
 | |
5WF1
 
 | |
6HYL
 
 | | Structure of PCM1 LIR motif bound to GABARAP | | Descriptor: | Pericentriolar material 1 protein,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HYO
 
 | | Structure of ULK1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HYM
 
 | | Structure of PCM1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pericentriolar material 1 protein,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HYN
 
 | | Structure of ATG13 LIR motif bound to GABARAP | | Descriptor: | Autophagy-related protein 13,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
4WN4
 
 | | Crystal structure of designed cPPR-polyA protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4WSL
 
 | | Crystal structure of designed cPPR-polyC protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
1G37
 
 | | CRYSTAL STRUCTURE OF HUMAN ALPHA-THROMBIN COMPLEXED WITH BCH-10556 AND EXOSITE-DIRECTED PEPTIDE | | Descriptor: | 3-(4-AMINO-CYCLOHEXYL)-2-HYDROXY-3-[(4-OXO-2-PHENYLMETHANESULFONYL-1,2,3,4-TETRAHYDRO-PYRROLO[1,2-A]PYRAZINE-6-CARBONYL)-AMINO]-PROPIONIC ACID BUTYL ESTER, ALPHA THROMBIN, THROMBIN NONAPEPTIDE INHIBITOR | | Authors: | Bachand, B, Tarazi, M, St-Denis, Y, Edmunds, J.J, Winocour, P.D, Leblond, L, Siddiqui, M.A. | | Deposit date: | 2000-10-23 | | Release date: | 2001-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and selective bicyclic lactam inhibitors of thrombin. Part 4: transition state inhibitors.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1IBD
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT V29A | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1IB5
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT W83Y | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-27 | | Release date: | 2001-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
4PJS
 
 | | Crystal structure of designed (SeMet)-cPPR-NRE protein | | Descriptor: | CALCIUM ION, Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJQ
 
 | | Crystal structure of designed cPPR-polyG protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
4PJR
 
 | | Crystal structure of designed cPPR-NRE protein | | Descriptor: | MAGNESIUM ION, Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
1IBH
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT M41I | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1IBB
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT W83F | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1IBF
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT V29G | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
