5ZRN
 
 | |
2GKO
 
 | | S41 Psychrophilic Protease | | Descriptor: | CALCIUM ION, SODIUM ION, microbial serine proteinases; subtilisin, ... | | Authors: | Walter, R.L, Mekel, M.J, Grayling, R.A, Arnold, F.H, Wintrode, P.L, Almog, O. | | Deposit date: | 2006-04-03 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state.
Proteins, 74, 2009
|
|
2HL0
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
2HL2
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with an analog of seryladenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
1H5N
 
 | | DMSO REDUCTASE MODIFIED BY THE PRESENCE OF DMS AND AIR | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM(VI) ION, ... | | Authors: | Bailey, S, Adams, B, Smith, A.T, Richards, R.L, Lowe, D.J, Bray, R.C. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reactions of Dimethyl Sulphoxide Reductase in the Presence of Dimethylsulphide and the Structure of the Dimethylsulphide-Modified Enzyme
Biochemistry, 40, 2001
|
|
2HKZ
 
 | |
2HL1
 
 | | Crystal structure of the editing domain of threonyl-tRNA synthetase from Pyrococcus abyssi in complex with seryl-3'-aminoadenosine | | Descriptor: | SERINE-3'-AMINOADENOSINE, Threonyl-tRNA synthetase | | Authors: | Hussain, T, Kruparani, S.P, Pal, B, Sankaranarayanan, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea
Embo J., 25, 2006
|
|
1Y2Q
 
 | |
4DQV
 
 | |
1E5V
 
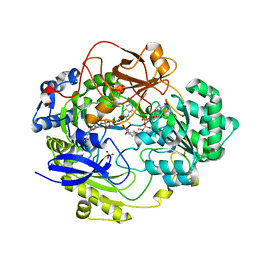 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
1E60
 
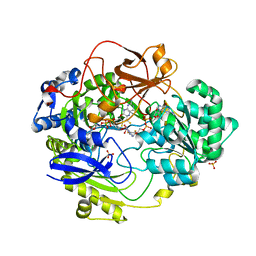 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
1E61
 
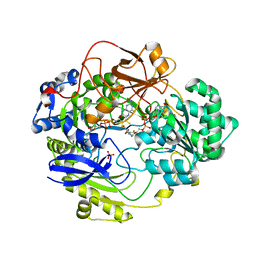 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
4RRQ
 
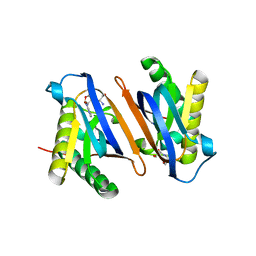 | |
5HT8
 
 | | Crystal structure of clostrillin double mutant (S17H,S19H) in complex with nickel | | Descriptor: | Beta and gamma crystallin, NICKEL (II) ION | | Authors: | Jamkhindikar, A, Srivastava, S.S, Sankaranarayanan, R. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Transition Metal-Binding, Trimeric beta gamma-Crystallin from Methane-Producing Thermophilic Archaea, Methanosaeta thermophila
Biochemistry, 56, 2017
|
|
5HT7
 
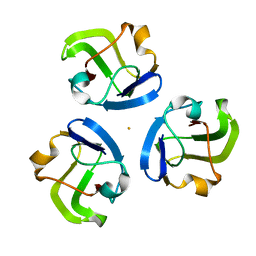 | |
4RRR
 
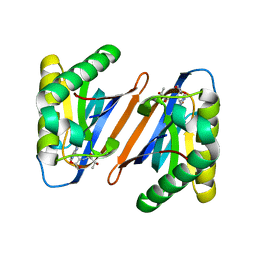 | |
1T9F
 
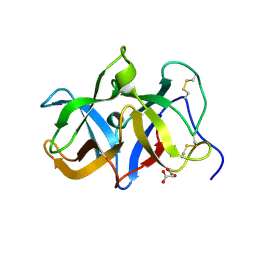 | | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function | | Descriptor: | MALONATE ION, protein 1d10 | | Authors: | Symersky, J, Li, S, Bunzel, R, Schormann, N, Luo, D, Huang, W.Y, Qiu, S, Gray, R, Zhang, Y, Arabashi, A, Lu, S, Luan, C.H, Tsao, J, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-16 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function.
To be Published
|
|
1T2N
 
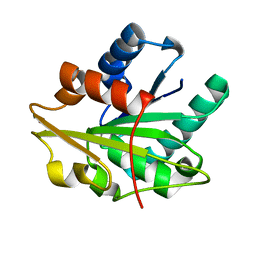 | |
2RN4
 
 | |
5H4E
 
 | |
5HT9
 
 | |
1U2Q
 
 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Weight Protein Tyrosine Phosphatase (MPtpA) at 2.5A resolution with glycerol in the active site | | Descriptor: | CHLORIDE ION, GLYCEROL, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
1U2P
 
 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Protein Tyrosine Phosphatase (MPtpA) at 1.9A resolution | | Descriptor: | CHLORIDE ION, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
1T4M
 
 | |
1SCZ
 
 | | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase | | Descriptor: | Dihydrolipoamide Succinyltransferase | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Tsao, J, Johnson, D, Huang, W.-Y, Pruett, P, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, Z, Lu, S, DeLucas, L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase
To be Published
|
|
