2RDU
 
 | |
2RDW
 
 | |
2RDT
 
 | | Crystal Structure of Human Glycolate Oxidase (GO) in Complex with CDST | | Descriptor: | 5-(dodecylthio)-1H-1,2,3-triazole-4-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Murray, M.S, Holmes, R.P, Lowther, W.T. | | Deposit date: | 2007-09-25 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Active Site and Loop 4 Movements within Human Glycolate Oxidase: Implications for Substrate Specificity and Drug Design.
Biochemistry, 47, 2008
|
|
6ULM
 
 | | Crystal structure of human cadherin 17 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin-17 | | Authors: | Gray, M.E, Sotomayor, M. | | Deposit date: | 2019-10-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the nonclassical cadherin-17 N-terminus and implications for its adhesive binding mechanism.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3NCL
 
 | |
3W80
 
 | |
1XW4
 
 | | Crystal Structure of Human Sulfiredoxin (Srx) in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Sulfiredoxin | | Authors: | Murray, M.S, Jonsson, T.J, Johnson, L.C, Poole, L.B, Lowther, W.T. | | Deposit date: | 2004-10-29 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin.
Biochemistry, 44, 2005
|
|
1XW3
 
 | | Crystal Structure of Human Sulfiredoxin (Srx) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, SULFIREDOXIN | | Authors: | Murray, M.S, Jonsson, T.J, Johnson, L.C, Poole, L.B, Lowther, W.T. | | Deposit date: | 2004-10-29 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin.
Biochemistry, 44, 2005
|
|
7DEV
 
 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora | | Descriptor: | Anthocyanin 5-aromatic acyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
7DEX
 
 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase H174A mutant with caffeoyl-CoA | | Descriptor: | Anthocyanin 5-aromatic acyltransferase, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (E)-3-[3,4-bis(oxidanyl)phenyl]prop-2-enethioate | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
3VTA
 
 | | Crystal Structure of cucumisin, a subtilisin-like endoprotease from Cucumis melo L | | Descriptor: | Cucumisin, DIISOPROPYL PHOSPHONATE, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Murayama, K, Kato-Murayama, M, Hosaka, T, Sotokawauchi, A, Shirouzu, M, Arima, K, Yokoyama, S. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of cucumisin, a subtilisin-like endoprotease from Cucumis melo L
J.Mol.Biol., 423, 2012
|
|
7YMQ
 
 | | Crystal structure of lysoplasmalogen specific phopholipase D, F211L mutant | | Descriptor: | Lysoplasmalogenase | | Authors: | Murayama, K, Kato-Murayama, M, Sugimori, D, Shirouzu, M, Hamana, H. | | Deposit date: | 2022-07-29 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
7YMP
 
 | | Crystal structure of lysoplasmalogen specific phospholipase D | | Descriptor: | Lysoplasmalogenase | | Authors: | Murayama, K, Kato-Murayama, M, Sugimori, D, Shirouzu, M, Hamana, H. | | Deposit date: | 2022-07-29 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
7YMR
 
 | | Complex structure of lysoplasmalogen specific phopholipase D, F211L mutant with LPC | | Descriptor: | Lysoplasmalogenase, [(2~{R})-2-oxidanyl-3-[oxidanyl-[2-(trimethyl-$l^{5}-azanyl)ethoxy]phosphoryl]oxy-propyl] hexadecanoate | | Authors: | Murayama, K, Kato-Murayama, M, Sugimori, D, Shirouzu, M, Hamana, H. | | Deposit date: | 2022-07-29 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
6MFT
 
 | | Crystal structure of glycosylated 426c HIV-1 gp120 core G459C in complex with glVRC01 A60C heavy chain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weidle, C, Pancera, M, Stamatatos, L, Gray, M. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
2DX1
 
 | | Crystal structure of RhoGEF protein Asef | | Descriptor: | Rho guanine nucleotide exchange factor 4 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-22 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of the rac activator, Asef, reveals its autoinhibitory mechanism
J.Biol.Chem., 282, 2007
|
|
3MDA
 
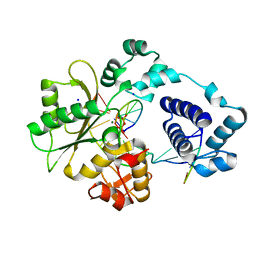 | | DNA polymerase lambda in complex with araC | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*CP*(CAR))-3'), DNA (5'-D(*CP*GP*GP*CP*GP*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Garcia-Diaz, M, Murray, M, Kunkel, T, Chou, K.M. | | Deposit date: | 2010-03-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Interaction between DNA Polymerase lambda and anticancer nucleoside analogs.
J.Biol.Chem., 285, 2010
|
|
2CYE
 
 | | Crystal structure of Thioesterase complexed with coenzyme A and Zn from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Putative thioesterase, ZINC ION | | Authors: | Hosaka, T, Kato-Murayama, M, Murayama, K, Kishishita, S, Handa, N, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2006-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the putative thioesterase protein TTHA1846 from Thermus thermophilus HB8 complexed with coenzyme A and a zinc ion.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2DXA
 
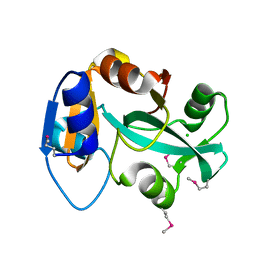 | | Crystal structure of trans editing enzyme ProX from E.coli | | Descriptor: | CHLORIDE ION, Protein ybaK | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of trans editing enzyme ProX from E.coli
To be Published
|
|
2DYC
 
 | | Crystal structure of the N-terminal domain of mouse galectin-4 | | Descriptor: | Galectin-4 | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-08 | | Release date: | 2007-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the N-terminal domain of mouse galectin-4
To be Published
|
|
2DXD
 
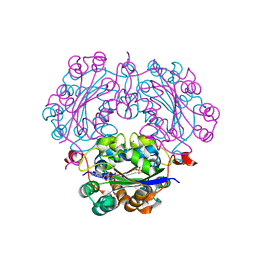 | | Crystal structure of nucleoside diphosphate kinase in complex with ATP analog | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Nucleoside diphosphate kinase | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with ATP analog
To be Published
|
|
2DYA
 
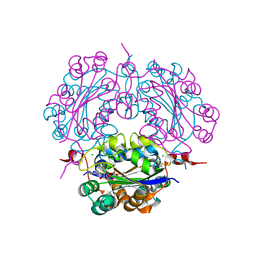 | | Crystal structure of complex between Adenine nucleotide and nucleoside diphosphate kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-08 | | Release date: | 2007-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of complex between Adenine nucleotide and nucleoside diphosphate kinase
To be Published
|
|
2DXE
 
 | | Crystal structure of nucleoside diphosphate kinase in complex with GDP | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with GDP
To be Published
|
|
2CWP
 
 | |
2CWK
 
 | |
