5OMG
 
 | | p38alpha in complex with pyrazolobenzothiazine inhibitor COXP4M12 | | Descriptor: | 3-(4-fluorophenyl)-4-methyl-1~{H}-pyrazolo[4,3-c][1,2]benzothiazine 5,5-dioxide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure determination and cellular evaluation of 1,4-dihydropyrazolo[4,3-c] [1,2] benzothiazine 5,5-dioxide p38 alpha MAPK inhibitors.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
3TZ9
 
 | | Kinase domain of cSrc in complex with RL130 | | Descriptor: | 1-[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]-3-[4-(quinazolin-4-ylamino)phenyl]urea, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gruetter, C, Richters, A, Rauh, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Overcoming Gatekeeper Mutations in cSrc and Abl by Hybrid Compound Design
To be Published
|
|
3TZ8
 
 | | Kinase domain of cSrc in complex with RL104 | | Descriptor: | N-(4-{[4-({[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]carbamoyl}amino)phenyl]amino}quinazolin-6-yl)-3-(4-methylpiperazin-1-yl)propanamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gruetter, C, Richters, A, Rauh, D. | | Deposit date: | 2011-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Overcoming Gatekeeper Mutations in cSrc and Abl by Hybrid Compound Design
To be Published
|
|
5OMH
 
 | | p38alpha in complex with pyrazolobenzothiazine inhibitor COXH11 | | Descriptor: | 1-(3-chlorophenyl)-3-methyl-4~{H}-pyrazolo[4,3-c][1,2]benzothiazine 5,5-dioxide, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Co-crystal structure determination and cellular evaluation of 1,4-dihydropyrazolo[4,3-c] [1,2] benzothiazine 5,5-dioxide p38 alpha MAPK inhibitors.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
2QLQ
 
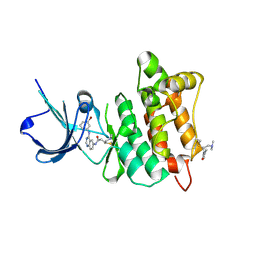 | | Crystal structure of SRC kinase domain with covalent inhibitor RL3 | | Descriptor: | (2E)-N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}-4-(dimethylamino)but-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Michalczyk, A, Rode, H.B, Gruetter, C, Rauh, D. | | Deposit date: | 2007-07-13 | | Release date: | 2008-03-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into how irreversible inhibitors can overcome drug resistance in EGFR.
Bioorg.Med.Chem., 16, 2008
|
|
2QI8
 
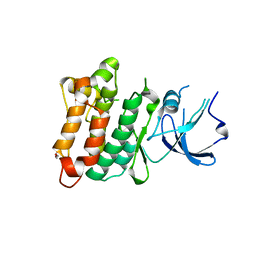 | | Crystal structure of drug resistant SRC kinase domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Michalczyk, A, Rode, H.B, Gruetter, C, Rauh, D. | | Deposit date: | 2007-07-03 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural insights into how irreversible inhibitors can overcome drug resistance in EGFR.
Bioorg.Med.Chem., 16, 2008
|
|
2QQ7
 
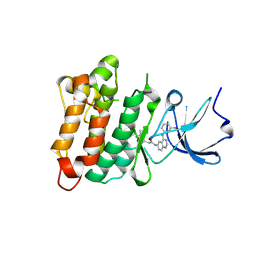 | | Crystal structure of drug resistant SRC kinase domain with irreversible inhibitor | | Descriptor: | (2E)-N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}-4-(dimethylamino)but-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Michalczyk, A, Rode, H.B, Gruetter, C, Rauh, D. | | Deposit date: | 2007-07-26 | | Release date: | 2008-03-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural insights into how irreversible inhibitors can overcome drug resistance in EGFR.
Bioorg.Med.Chem., 16, 2008
|
|
8C72
 
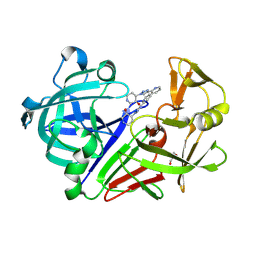 | | Pyrrolidine fragment 10b bound to endothiapepsin | | Descriptor: | (3~{S},4~{S})-4-(4-pyridin-2-yl-1,2,3-triazol-1-yl)piperidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C71
 
 | | Pyrrolidine fragment 5b bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-(3,4-dihydro-1~{H}-isoquinolin-2-yl)pyrrolidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C74
 
 | | Pyrrolidine fragment 10d bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-[4-[(4-azanylphenoxy)methyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6P
 
 | | Fragment screening hit I bound to endothiapepsin | | Descriptor: | 4-[(2-azanyl-4-methyl-1,3-thiazol-5-yl)methyl]benzenecarbonitrile, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6T
 
 | | Fragment screening hit IV bound to endothiapepsin | | Descriptor: | 1-[3,5-bis(chloranyl)phenoxy]propan-2-amine, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6S
 
 | | Fragment screening hit III bound to endothiapepsin | | Descriptor: | 4-(1,4-diazepan-1-ylsulfonyl)isoquinoline, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6Q
 
 | | Fragment screening hit II bound to endothiapepsin | | Descriptor: | 3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)aniline, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C70
 
 | | Pyrrolidine fragment 1 bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-[4-[(5-bromanylpyridin-3-yl)oxymethyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
6GN1
 
 | | Crystal Structure of Glycogen synthase kinase-3 beta (GSK3B) in Complex with PIK-75 | | Descriptor: | CHLORIDE ION, Glycogen synthase kinase-3 beta, ~{N}-[(~{E})-(6-bromanylimidazo[1,2-a]pyridin-3-yl)methylideneamino]-~{N},2-dimethyl-5-nitro-benzenesulfonamide | | Authors: | Tesch, R, Becker, C, Mueller, M.P, Sant'Anna, C.M.R, Fraga, C.A.M, Rauh, D. | | Deposit date: | 2018-05-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Unusual Intramolecular Halogen Bond Guides Conformational Selection.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7ZYN
 
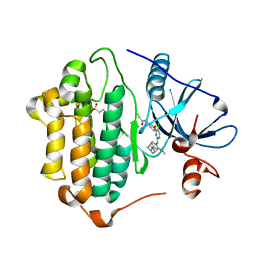 | | Crystal Structure of EGFR-T790M/C797S in Complex with WZ4002 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, N-{3-[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
7ZYP
 
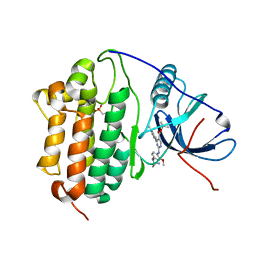 | | Crystal Structure of EGFR-T790M/C797S in Complex with Reversible Aminopyrimidine 9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, propan-2-yl 2-[[4-(4-azanylpiperidin-1-yl)-2-methoxy-phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
4L8M
 
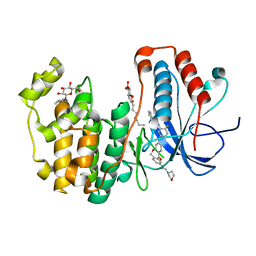 | | Human p38 MAP kinase in complex with a Dibenzoxepinone | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-fluoro-5-({9-[2-(morpholin-4-yl)ethoxy]-11-oxo-6,11-dihydrodibenzo[b,e]oxepin-3-yl}amino)phenyl]benzamide, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2013-06-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metabolically Stable Dibenzo[b,e]oxepin-11(6H)-ones as Highly Selective p38 MAP Kinase Inhibitors: Optimizing Anti-Cytokine Activity in Human Whole Blood.
J.Med.Chem., 56, 2013
|
|
7ZYM
 
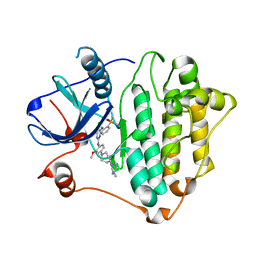 | | Crystal Structure of EGFR-T790M/C797S in Complex with Brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
6Z4B
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with Osimertinib and EAI045 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, SULFATE ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complex Crystal Structures of EGFR with Third-Generation Kinase Inhibitors and Simultaneously Bound Allosteric Ligands.
Acs Med.Chem.Lett., 11, 2020
|
|
6Z4D
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with Mavelertinib and EAI001 | | Descriptor: | (2R)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-2-phenyl-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex Crystal Structures of EGFR with Third-Generation Kinase Inhibitors and Simultaneously Bound Allosteric Ligands.
Acs Med.Chem.Lett., 11, 2020
|
|
7ZYQ
 
 | | Crystal Structure of EGFR-T790M/V948R in Complex with Reversible Aminopyrimidine 13 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, SODIUM ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
7NH5
 
 | |
7NH4
 
 | |
