4MZD
 
 | | High resolution crystal structure of the nisin leader peptidase NisP from Lactococcus lactis | | Descriptor: | Nisin leader peptide-processing serine protease NisP | | Authors: | Rao, Z.H, Xu, Y.Y, Li, X, Yang, W. | | Deposit date: | 2013-09-30 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the nisin leader peptidase NisP revealing a C-terminal autocleavage activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5YK1
 
 | | The complex structure of Rv3197-AMPPNP from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable conserved ATP-binding protein ABC transporter, ... | | Authors: | Rao, Z.H, Zhang, Q.Q. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
5YK2
 
 | |
5YK0
 
 | | The complex structure of Rv3197-ADP from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Probable conserved ATP-binding protein ABC transporter, ... | | Authors: | Rao, Z.H, Zhang, Q.Q. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
3FFM
 
 | |
6M5S
 
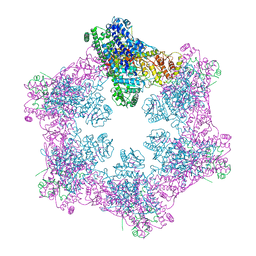 | | The coordinates of the apo hexameric terminase complex | | Descriptor: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5T
 
 | | The coordinate of the nuclease domain of the apo terminase complex | | Descriptor: | Tripartite terminase subunit 3 | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5R
 
 | | The coordinates of the apo monomeric terminase complex | | Descriptor: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5V
 
 | | The coordinate of the hexameric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M5U
 
 | | The coordinates of the monomeric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Zhu, L, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
4R3D
 
 | | Crystal structure of MERS Coronavirus papain like protease | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Kong, L.Y, Wang, Q, Ming, Z.H, Shaw, N, Sun, Y.N, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | The Crystal Structures of Middle East Respiratory Syndrome Coronavirus Papain-like Protease and Its Complex with an Inhibitor
To be Published
|
|
5HYW
 
 | | The crystal structure of the D3-ASK1 complex | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
5HZG
 
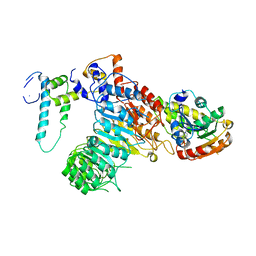 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | Descriptor: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | Authors: | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | Deposit date: | 2016-02-02 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
4K7E
 
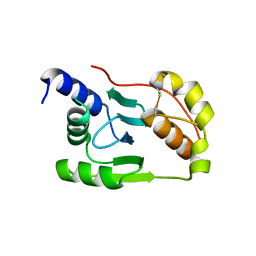 | | Crystal structure of Junin virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Zhang, Y.J, Li, L, Liu, X, Dong, S.S, Wang, W.M, Huo, T, Rao, Z.H, Yang, C. | | Deposit date: | 2013-04-17 | | Release date: | 2013-08-07 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Junin virus nucleoprotein
J.Gen.Virol., 94, 2013
|
|
8YMK
 
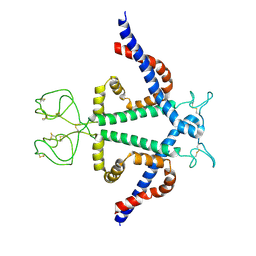 | | Localized reconstruction of Hepatitis B virus surface antigen dimer in the subviral particle with D2 symmetry from dataset A0 | | Descriptor: | Isoform S of Large envelope protein | | Authors: | Wang, T, Cao, L, Mu, A, Wang, Q, Rao, Z.H. | | Deposit date: | 2024-03-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inherent symmetry and flexibility in hepatitis B virus subviral particles.
Science, 385, 2024
|
|
8YMJ
 
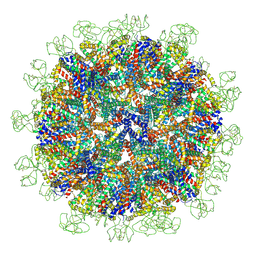 | | Cryo-EM structure of Hepatitis B virus surface antigen subviral particle with D2 symmetry | | Descriptor: | Isoform S of Large envelope protein | | Authors: | Wang, T, Cao, L, Mu, A, Wang, Q, Rao, Z.H. | | Deposit date: | 2024-03-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Inherent symmetry and flexibility in hepatitis B virus subviral particles.
Science, 385, 2024
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6D
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | Descriptor: | Infectious bronchitis virus (IBV) main protease | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | Descriptor: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
1J4G
 
 | | crystal structure analysis of the trichosanthin delta C7 | | Descriptor: | CALCIUM ION, Trichosanthin delta C7 | | Authors: | Ding, Y, Too, H.M, Wang, Z.L, Liu, Y.W, Dong, Y.C, Shaw, P.C, Rao, Z.H. | | Deposit date: | 2001-09-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure analysis of the trichosanthin delta C7
To be Published
|
|
4Z9P
 
 | | Crystal structure of Ebola virus nucleoprotein core domain at 1.8A resolution | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Dong, S.S, Yang, P, Li, G.B, Liu, B.C, Yang, C, Rao, Z.H. | | Deposit date: | 2015-04-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution.
Protein Cell, 6, 2015
|
|
1J0W
 
 | |
1LSP
 
 | | THE CRYSTAL STRUCTURE OF A BULGECIN-INHIBITED G-TYPE LYSOZYME FROM THE EGG-WHITE OF THE AUSTRALIAN BLACK SWAN. A COMPARISON OF THE BINDING OF BULGECIN TO THREE MURAMIDASES | | Descriptor: | BULGECIN A, LYSOZYME | | Authors: | Karlsen, S, Rao, Z.H, Hough, E, Isaacs, N.W. | | Deposit date: | 1995-02-09 | | Release date: | 1996-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of a bulgecin-inhibited g-type lysozyme from the egg white of the Australian black swan. A comparison of the binding of bulgecin to three muramidases.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
5GJB
 
 | | Zika virus NS3 helicase in complex with ssRNA | | Descriptor: | NS3 helicase, RNA (5'-R(*AP*GP*AP*UP*CP*AP*A)-3') | | Authors: | Tian, H.L, Ji, X.Y, Yang, X.Y, Zhang, Z.X, Lu, Z.K, Yang, K.L, Chen, C, Zhao, Q, Chi, H, Mu, Z.Y, Xie, W, Wang, Z.F, Lou, H.Q, Yang, H.T, Rao, Z.H. | | Deposit date: | 2016-06-28 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis of Zika virus helicase in recognizing its substrates
Protein Cell, 7, 2016
|
|
