2M4F
 
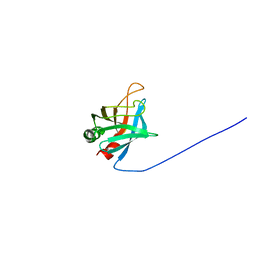 | | Solution Structure of Outer surface protein E | | Descriptor: | Outer surface protein E | | Authors: | Bhattacharjee, A, Oeemig, J.S, Kolodziejczyk, R, Meri, T, Kajander, T, Iwai, H, Jokiranta, T, Goldman, A. | | Deposit date: | 2013-02-05 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Complement Evasion by Lyme Disease Pathogen Borrelia burgdorferi
J.Biol.Chem., 288, 2013
|
|
6N1R
 
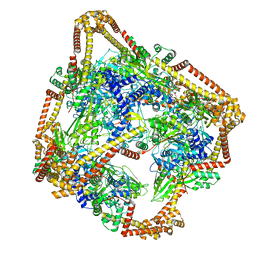 | | Tetrahedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in tetrahedral symmetry | | Descriptor: | DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
1FD8
 
 | | SOLUTION STRUCTURE OF THE CU(I) FORM OF THE YEAST METALLOCHAPERONE, ATX1 | | Descriptor: | ATX1 COPPER CHAPERONE, COPPER (I) ION | | Authors: | Arnesano, F, Banci, L, Bertini, I, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-07-20 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1.
Biochemistry, 40, 2001
|
|
1FES
 
 | | SOLUTION STRUCTURE OF THE APO FORM OF THE YEAST METALLOCHAPERONE, ATX1 | | Descriptor: | ATX1 COPPER CHAPERONE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-07-22 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1.
Biochemistry, 40, 2001
|
|
6N1P
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment with DNA, solved by cryoEM in C2 symmetry | | Descriptor: | DNA (44-MER), DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.35 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
7P0D
 
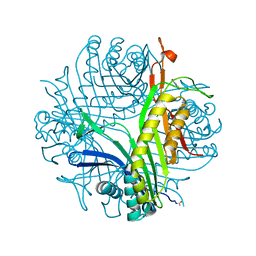 | |
7P0G
 
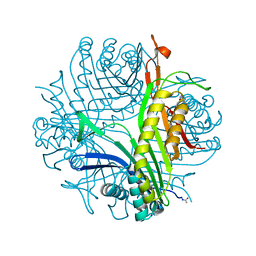 | |
4WGG
 
 | | STRUCTURE OF THE TERNARY COMPLEX OF A ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE IN COMPLEX WITH NADP AND CONIFERYL ALDEHYDE | | Descriptor: | (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enal, Double Bond Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Collery, J, Langlois d'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2014-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | STRUCTURE OF ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE
to be published
|
|
7P0C
 
 | |
5NSW
 
 | | Xenon for tunnelling analysis of the efflux pump component OprN. | | Descriptor: | Multidrug efflux outer membrane protein OprN, NICKEL (II) ION, PALMITIC ACID, ... | | Authors: | Phan, G, Prange, T, Enguene Ntsogo, Y.V, Garnier, C, Ducruix, A, Broutin, I. | | Deposit date: | 2017-04-27 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenon for tunnelling analysis of the efflux pump component OprN.
PLoS ONE, 12, 2017
|
|
6N1Q
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in D2 symmetry | | Descriptor: | DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
1H9W
 
 | | Native Dioclea Guianensis seed lectin | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SEED LECTIN | | Authors: | Romero, A, Wah, D.A, Sol, F.G.D, Cavada, B.S, Ramos, M.V, Grangeiro, T.B, Sampaio, A.H, Calvete, J.J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Native and Cd/Cd-Substituted Dioclea Guianensis Seed Lectin. A Novel Manganese-Binding Site and Structural Basis of Dimer-Tetramer Association
J.Mol.Biol., 310, 2001
|
|
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
4WOG
 
 | | Crystal Structure of Frutalin from Artocarpus incisa | | Descriptor: | Frutalin | | Authors: | Pereira, H.M, Moreira, A.C.O.M, Vieira Neto, A.E, Moreno, F.B.M.B, Lobo, M.D.P, Sousa, F.D, Grangeiro, T.B, Moreira, R.A. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Crystal Structure of Frutalin from Artocarpus incisa
To Be Published
|
|
4WLS
 
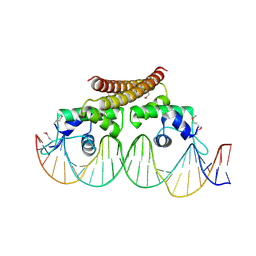 | | Crystal structure of the metal-free (repressor) form of E. Coli CUER, a copper efflux regulator, bound to COPA promoter DNA | | Descriptor: | COPA PROMOTER DNA NON-TEMPLATE STRAND, COPA PROMOTER DNA NON-TEMPLATE STRAND (ALTERNATE CONFORMATION), COPA PROMOTER DNA TEMPLATE STRAND, ... | | Authors: | Philips, S.J, Canalizo-Hernandez, M, Mondragon, A, O'Halloran, T.V. | | Deposit date: | 2014-10-08 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Allosteric transcriptional regulation via changes in the overall topology of the core promoter.
Science, 349, 2015
|
|
1H9P
 
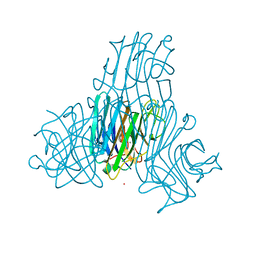 | | Crystal Structure of Dioclea guianensis Seed Lectin | | Descriptor: | CADMIUM ION, LECTIN ALPHA CHAIN, MANGANESE (II) ION | | Authors: | Romero, A, Wah, D.A, Gallego Del sol, F, Cavada, B.S, Ramos, M.V, Grangeiro, T.B, Sampaio, A.H, Calvete, J.J. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Native and Cd/Cd-Substituted Dioclea Guianensis Seed Lectin. A Novel Manganese-Binding Site and Structural Basis of Dimer-Tetramer Association
J.Mol.Biol., 310, 2001
|
|
6OU9
 
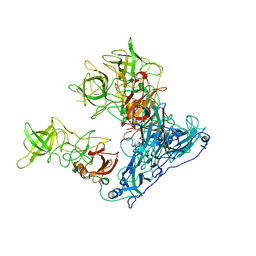 | | Asymmetric focused reconstruction of human norovirus GI.7 Houston strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Major capsid protein | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-04 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OUT
 
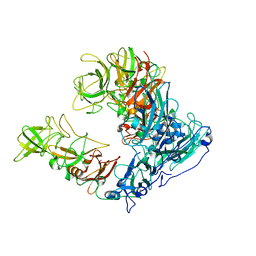 | | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Capsid protein VP1 | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5NII
 
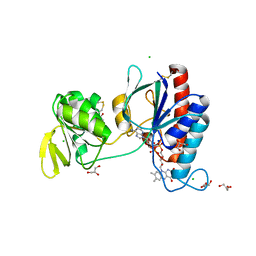 | | Crystal structure of the atypical thioredoxin reductase TRi from Desulfovibrio vulgaris Hildenborough | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Valette, O, Tran, T.T.I, Cavazza, C, Caudeville, E, Brasseur, G, Dolla, A, Talla, E, Pieulle, L. | | Deposit date: | 2017-03-24 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical Function, Molecular Structure and Evolution of an Atypical Thioredoxin Reductase from Desulfovibrio vulgaris.
Front Microbiol, 8, 2017
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
5LO9
 
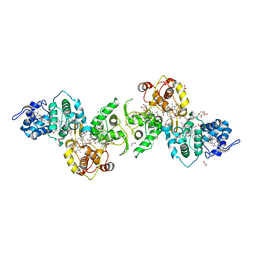 | | Thiosulfate dehydrogenase (TsdBA) from Marichromatium purpuratum - "as isolated" form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytochrome C, ... | | Authors: | Brito, J.A, Kurth, J.M, Reuter, J, Flegler, A, Koch, T, Franke, T, Klein, E, Rowe, S, Butt, J.N, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2016-08-08 | | Release date: | 2016-10-12 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Electron Accepting Units of the Diheme Cytochrome c TsdA, a Bifunctional Thiosulfate Dehydrogenase/Tetrathionate Reductase.
J.Biol.Chem., 291, 2016
|
|
6OUC
 
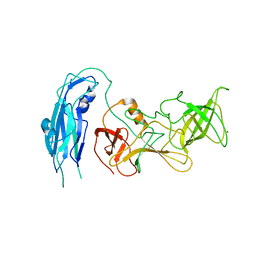 | | Asymmetric focsued reconstruction of human norovirus GII.2 Snow Mountain Virus strain VLP asymmetric unit in T=1 symmetry | | Descriptor: | Viral protein 1, ZINC ION | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-04 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4YVB
 
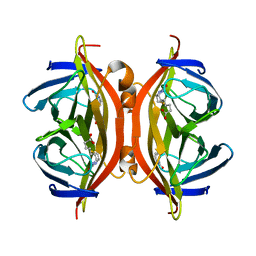 | | Structure of D128N streptavidin | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Baugh, L, Le Trong, I, Stayton, P.S, Stenkamp, R.E, Lybrand, T.P. | | Deposit date: | 2015-03-19 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | A Streptavidin Binding Site Mutation Yields an Unexpected Result
To Be Published
|
|
6OUU
 
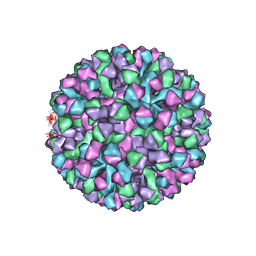 | | Symmetric reconstruction of human norovirus GII.4 Minerva strain VLP in T=4 symmetry | | Descriptor: | Major capsid protein | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
