4C6D
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 6.0 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6F
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 6.5 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, DIHYDROOROTASE, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4BY3
 
 | | Crystal structure of the dihydroorotase domain of human CAD in apo- form obtained recombinantly from E. coli. | | Descriptor: | DIHYDROOROTASE, FORMIC ACID, ZINC ION | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-07-17 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure, Functional Characterization, and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6L
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to the inhibitor fluoroorotate at pH 6.0 | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6B
 
 | | Crystal structure of the dihydroorotase domain of human CAD with incomplete active site, obtained recombinantly from E. coli. | | Descriptor: | CAD PROTEIN, FORMIC ACID, GLYCEROL | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.656 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6P
 
 | | Crystal structure of the dihydroorotase domain of human CAD C1613S mutant in apo-form at pH 7.0 | | Descriptor: | CAD PROTEIN, FORMIC ACID, ZINC ION | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.518 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6Q
 
 | | Crystal structure of the dihydroorotase domain of human CAD C1613S mutant bound to substrate at pH 7.0 | | Descriptor: | CAD PROTEIN, FORMIC ACID, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6J
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 7.5 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
6RNS
 
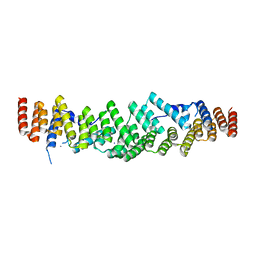 | |
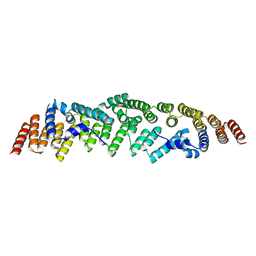
6RNQ
 
 | |
3MR6
 
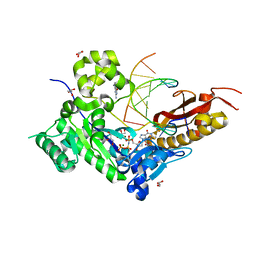 | | Human DNA polymerase eta - DNA ternary complex with a CPD 2bp upstream of the active site (TT4) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*C*AP*TP*CP*AP*(TTD)P*AP*CP*GP*AP*GP*C)-3'), DNA (5'-D(*TP*CP*TP*CP*GP*TP*AP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3MR2
 
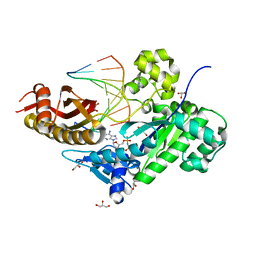 | | Human DNA polymerase eta in complex with normal DNA and incoming nucleotide (Nrm) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
3SI8
 
 | | Human DNA polymerase eta - DNA ternary complex with the 5'T of a CPD in the active site (TT2) | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, ... | | Authors: | Biertumpfel, C, Zhao, Y, Kondo, Y, Ramon-Maiques, S, Gregory, M, Lee, J.Y, Masutani, C, Lehmann, A.R, Hanaoka, F, Yang, W. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
1X9Z
 
 | | Crystal structure of the MutL C-terminal domain | | Descriptor: | CHLORIDE ION, DNA mismatch repair protein mutL, GLYCEROL, ... | | Authors: | Guarne, A, Ramon-Maiques, S, Wolff, E.M, Ghirlando, R, Hu, X, Miller, J.H, Yang, W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair
Embo J., 23, 2004
|
|
2MQK
 
 | |
5G1N
 
 | | Aspartate transcarbamoylase domain of human CAD bound to PALA | | Descriptor: | 1,2-ETHANEDIOL, CAD PROTEIN, N-(PHOSPHONACETYL)-L-ASPARTIC ACID | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
5G1O
 
 | | Aspartate transcarbamoylase domain of human CAD in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAD protein, GLYCEROL | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
3MR5
 
 | | Human DNA polymerase eta - DNA ternary complex with a CPD 1bp upstream of the active site (TT3) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*CP*GP*TP*CP*AP*TP*AP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*(TTD)P*AP*TP*GP*AP*CP*GP*A)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
5G1P
 
 | | Aspartate transcarbamoylase domain of human CAD bound to carbamoyl phosphate | | Descriptor: | CAD PROTEIN, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
3MR3
 
 | | Human DNA polymerase eta - DNA ternary complex with the 3'T of a CPD in the active site (TT1) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*AP*(TTD)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
8BPS
 
 | |
8BPL
 
 | |
5NNQ
 
 | |
5NNN
 
 | |
4BS1
 
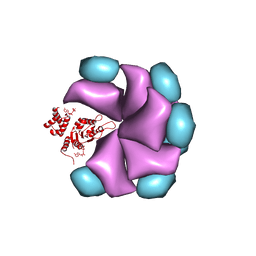 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR (NTRC FAMILY) | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
