6LVR
 
 | |
9L6Y
 
 | | Crystal structure of the L7Ae derivative protein LS12 in complex with its co-evolved target CS2 RNA | | Descriptor: | CALCIUM ION, CS2 RNA, Large ribosomal subunit protein eL8 | | Authors: | Teramoto, T, Nakashima, M, Fukunaga, K, Yokobayashi, Y, Kakuta, Y. | | Deposit date: | 2024-12-25 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural insights into lab-coevolved RNA-RBP pairs and applications of synthetic riboswitches in cell-free system.
Nucleic Acids Res., 53, 2025
|
|
9L6X
 
 | | Crystal structure of the L7Ae derivative protein LS4 in complex with its co-evolved target CS1 RNA | | Descriptor: | CS1 RNA, Large ribosomal subunit protein eL8, MAGNESIUM ION | | Authors: | Teramoto, T, Nakashima, M, Fukunaga, K, Yokobayashi, Y, Kakuta, Y. | | Deposit date: | 2024-12-25 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into lab-coevolved RNA-RBP pairs and applications of synthetic riboswitches in cell-free system.
Nucleic Acids Res., 53, 2025
|
|
9L6Z
 
 | | Crystal structure of the L7Ae derivative protein LS12 in RNA-free state | | Descriptor: | Large ribosomal subunit protein eL8, SODIUM ION | | Authors: | Teramoto, T, Nakashima, M, Fukunaga, K, Yokobayashi, Y, Kakuta, Y. | | Deposit date: | 2024-12-25 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into lab-coevolved RNA-RBP pairs and applications of synthetic riboswitches in cell-free system.
Nucleic Acids Res., 53, 2025
|
|
7F3E
 
 | | Cryo-EM structure of the minimal protein-only RNase P from Aquifex aeolicus | | Descriptor: | RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Adachi, N, Kawasaki, M, Moriya, T, Numata, T, Senda, T, Kakuta, Y. | | Deposit date: | 2021-06-16 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Minimal protein-only RNase P structure reveals insights into tRNA precursor recognition and catalysis.
J.Biol.Chem., 297, 2021
|
|
5UZG
 
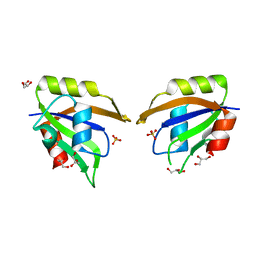 | | Crystal structure of Glorund qRRM1 domain | | Descriptor: | AT27789p, GLYCEROL, SULFATE ION | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
5UZM
 
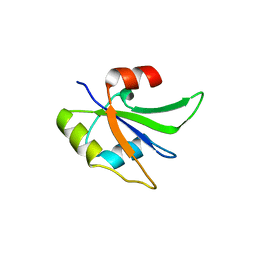 | | Crystal structure of Glorund qRRM2 domain | | Descriptor: | AT27789p | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
7EOV
 
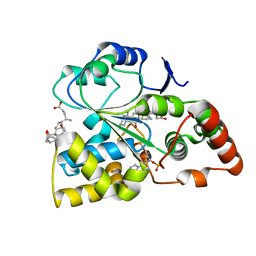 | | Crystal structure of mouse cytosolic sulfotransferase mSULT2A8 in complex with PAP and cholic acid | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CHOLIC ACID, cytosolic sulfotransferase SULT2A8 | | Authors: | Teramoto, T, Nishio, T, Kakuta, Y. | | Deposit date: | 2021-04-22 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of mouse SULT2A8 reveals the mechanism of 7 alpha-hydroxyl, bile acid sulfation.
Biochem.Biophys.Res.Commun., 562, 2021
|
|
5UZN
 
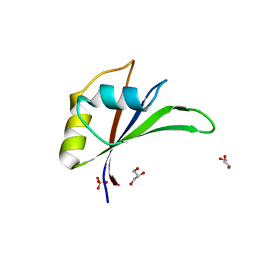 | | Crystal structure of Glorund qRRM3 domain | | Descriptor: | AT27789p, GLYCEROL, SULFATE ION | | Authors: | Teramoto, T, Hall, T.M.T. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Drosophila hnRNP F/H Homolog Glorund Uses Two Distinct RNA-Binding Modes to Diversify Target Recognition.
Cell Rep, 19, 2017
|
|
7V1O
 
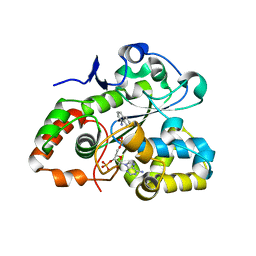 | |
3AP2
 
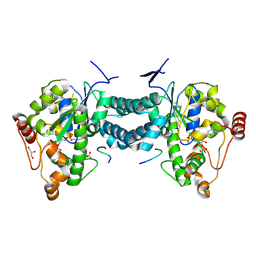 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP,C4 peptide, and phosphate ion | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2: Insights into substrate-binding and catalysis of post-translational protein tyrosine sulfation
To be Published
|
|
3AP1
 
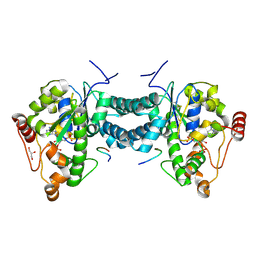 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP and C4 peptide | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
2ZVP
 
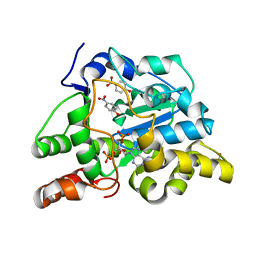 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAP and p-nitrophenol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the broad range substrate specificity of a novel mouse cytosolic sulfotransferase--mSULT1D1
Biochem.Biophys.Res.Commun., 379, 2009
|
|
2ZYU
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAPS and p-nitrophenyl sulfate | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, 4-nitrophenyl sulfate, GLYCEROL, ... | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshot of a Michaelis complex in a sulfuryl transfer reaction: Crystal structure of a mouse sulfotransferase, mSULT1D1, complexed with donor substrate and accepter substrate
Biochem.Biophys.Res.Commun., 383, 2009
|
|
2ZYV
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAPS/PAP and p-nitrophenol | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Snapshot of a Michaelis complex in a sulfuryl transfer reaction: Crystal structure of a mouse sulfotransferase, mSULT1D1, complexed with donor substrate and accepter substrate
Biochem.Biophys.Res.Commun., 383, 2009
|
|
2ZYW
 
 | | crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAP and p-nitrophenol, obtained by two-step soaking method | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshot of a Michaelis complex in a sulfuryl transfer reaction: Crystal structure of a mouse sulfotransferase, mSULT1D1, complexed with donor substrate and accepter substrate
Biochem.Biophys.Res.Commun., 383, 2009
|
|
2ZYT
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAPS | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, GLYCEROL, Tyrosine-ester sulfotransferase | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2009-01-29 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshot of a Michaelis complex in a sulfuryl transfer reaction: Crystal structure of a mouse sulfotransferase, mSULT1D1, complexed with donor substrate and accepter substrate
Biochem.Biophys.Res.Commun., 383, 2009
|
|
2ZPT
 
 | | Crystal structure of mouse sulfotransferase SULT1D1 complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, Tyrosine-ester sulfotransferase | | Authors: | Teramoto, T, Sakakibara, Y, Inada, K, Liu, M.C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2008-07-28 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of mSULT1D1, a mouse catecholamine sulfotransferase
Febs Lett., 582, 2008
|
|
2ZVQ
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT1D1 complex with PAP and alpha-naphthol | | Descriptor: | 1-NAPHTHOL, ADENOSINE-3'-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Teramoto, T, Sakakibara, Y, Liu, M.-C, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the broad range substrate specificity of a novel mouse cytosolic sulfotransferase--mSULT1D1
Biochem.Biophys.Res.Commun., 379, 2009
|
|
3AP3
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Protein-tyrosine sulfotransferase 2 | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
1Z2T
 
 | | NMR structure study of anchor peptide Ser65-Leu87 of enzyme acholeplasma laidlawii Monoglycosyldiacyl Glycerol Synthase (alMGS) in DHPC micelles | | Descriptor: | Anchor peptide Ser65-Leu87 of alMGS | | Authors: | Lind, J, Barany-Wallje, E, Ramo, T, Wieslander, A, Maler, L. | | Deposit date: | 2005-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, position of and membrane-interaction of a putative membrane-anchoring domain of alMGS
To be Published
|
|
7YQ1
 
 | | Crystal structure of adenosine 5'-phosphosulfate kinase from Archaeoglobus fulgidus in complex with AMP-PNP and APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, Adenylyl-sulfate kinase, BORIC ACID, ... | | Authors: | Kawakami, T, Teramoto, T, Kakuta, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of adenosine 5'-phosphosulfate kinase isolated from Archaeoglobus fulgidus.
Biochem.Biophys.Res.Commun., 643, 2022
|
|
7YRO
 
 | | Crystal structure of mango fucosyltransferase 13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Fucosyltransferase, ... | | Authors: | Okada, T, Teramoto, T, Ihara, H, Ikeda, Y, Kakuta, Y. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of mango alpha 1,3/ alpha 1,4-fucosyltransferase elucidates unique elements that regulate Lewis A-dominant oligosaccharide assembly.
Glycobiology, 34, 2024
|
|
8J12
 
 | | Cryo-EM structure of the AsCas12f-sgRNA-target DNA ternary complex | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (247-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-12 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
8J1J
 
 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
