2E82
 
 | | Crystal structure of human D-amino acid oxidase complexed with imino-DOPA | | Descriptor: | (2E)-3-(3,4-DIHYDROXYPHENYL)-2-IMINOPROPANOIC ACID, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kawazoe, T, Tsuge, H, Imagawa, T, Kuramitsu, S, Fukui, K. | | Deposit date: | 2007-01-16 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of d-DOPA oxidation by d-amino acid oxidase: Alternative pathway for dopamine biosynthesis.
Biochem.Biophys.Res.Commun., 355, 2007
|
|
2ZQE
 
 | | Crystal structure of the Smr domain of Thermus thermophilus MutS2 | | Descriptor: | MutS2 protein | | Authors: | Fukui, K, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2008-08-08 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MutS2 endonuclease domain and the mechanism of homologous recombination suppression
J.Biol.Chem., 283, 2008
|
|
2WQK
 
 | | Crystal Structure of Sure Protein from Aquifex aeolicus | | Descriptor: | 5'-NUCLEOTIDASE SURE, SODIUM ION, SULFATE ION | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-08-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Sure Protein from Aquifex Aeolicus Vf5 at 1.5 A Resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2D64
 
 | | Aspartate Aminotransferase Mutant MABC With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
3AB7
 
 | |
4LQ6
 
 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3AKD
 
 | |
3ANG
 
 | | Crystal structure of Thermus thermophilus FadR in complex with E. coli-derived dodecyl-CoA | | Descriptor: | DODECYL-COA, Transcriptional repressor, TetR family | | Authors: | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2010-09-01 | | Release date: | 2011-03-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation.
Microbiology, 157, 2011
|
|
3ANP
 
 | | Crystal structure of Thermus thermophilus FadR, a TetR familly transcriptional repressor, in complex with lauroyl-CoA. | | Descriptor: | DODECYL-COA, LAURIC ACID, Transcriptional repressor, ... | | Authors: | Agari, Y, Agari, K, Sakamoto, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2010-09-06 | | Release date: | 2011-03-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation.
Microbiology, 157, 2011
|
|
3ASZ
 
 | | CMP-complex structure of uridine kinase from Thermus thermophilus HB8 | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Uridine kinase | | Authors: | Tomoike, F, Nakagawa, N, Kuramitsu, S, Masui, R. | | Deposit date: | 2010-12-22 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Single Amino Acid Limits the Substrate Specificity of Thermus thermophilus Uridine-Cytidine Kinase to Cytidine
Biochemistry, 50, 2011
|
|
3ASY
 
 | |
3ADR
 
 | | The first crystal structure of an archaeal metallo-beta-lactamase superfamily protein; ST1585 from Sulfolobus tokodaii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative uncharacterized protein ST1585, ... | | Authors: | Shimada, A, Ishikawa, H, Nakagawa, N, Kuramitsu, S, Masui, R. | | Deposit date: | 2010-01-28 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first crystal structure of an archaeal metallo-beta-lactamase superfamily protein; ST1585 from Sulfolobus tokodaii
Proteins, 78, 2010
|
|
3AKC
 
 | | Crystal structure of CMP kinase in complex with CDP and ADP from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, Cytidylate kinase, ... | | Authors: | Mega, R, Nakagawa, N, Kuramitsu, S, Masui, R. | | Deposit date: | 2010-07-12 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of the tertiary complex of CMP kinase with a phosphoryl group acceptor and a donor from Thermus thermophilus HB8
To be Published
|
|
2ZXR
 
 | | Crystal structure of RecJ in complex with Mg2+ from Thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, Single-stranded DNA specific exonuclease RecJ | | Authors: | Wakamatsu, T, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of RecJ exonuclease defines its specificity for single-stranded DNA
J.Biol.Chem., 285, 2010
|
|
3AAI
 
 | |
3AB8
 
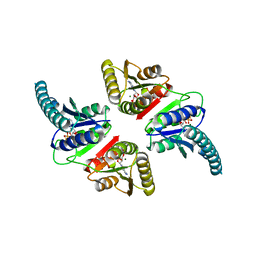 | |
3B02
 
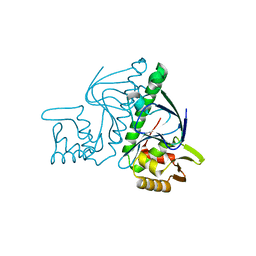 | | Crystal structure of TTHB099, a transcriptional regulator CRP family from Thermus thermophilus HB8 | | Descriptor: | Transcriptional regulator, Crp family | | Authors: | Agari, Y, Kuramitsu, S, Shinkai, A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | X-ray crystal structure of TTHB099, a CRP/FNR superfamily transcriptional regulator from Thermus thermophilus HB8, reveals a DNA-binding protein with no required allosteric effector molecule
Proteins, 80, 2012
|
|
1TFM
 
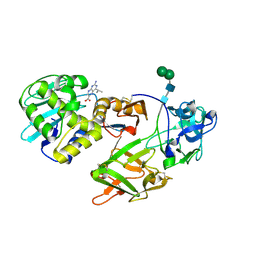 | | CRYSTAL STRUCTURE OF A RIBOSOME INACTIVATING PROTEIN IN ITS NATURALLY INHIBITED FORM | | Descriptor: | 2-AMINO-4-ISOPROPYL-PTERIDINE-6-CARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mishra, V, Bilgrami, S, Paramasivam, M, Yadav, S, Sharma, R.S, Kaur, P, Srinivasan, A, Babu, C.R, Singh, T.P. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF A RIBOSOME INACTIVATING PROTEIN IN ITS NATURALLY INHIBITED FORM
To be Published
|
|
4OJA
 
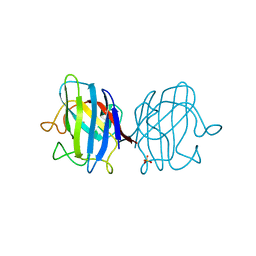 | |
4QDM
 
 | | Crystal structure of N-terminal mutant (V1L) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Alkaline thermostable endoxylanase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2014-05-14 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Structural insights into N-terminal to C-terminal interactions and implications for thermostability of a (beta/alpha)8-triosephosphate isomerase barrel enzyme
Febs J., 282, 2015
|
|
4QCE
 
 | | Crystal structure of recombinant alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Alkaline thermostable endoxylanase, MAGNESIUM ION, SODIUM ION | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2014-05-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural insights into N-terminal to C-terminal interactions and implications for thermostability of a (beta/alpha)8-triosephosphate isomerase barrel enzyme
Febs J., 282, 2015
|
|
4QE8
 
 | | FXR with DM175 and NCoA-2 peptide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 4-({2-[(4-tert-butylbenzoyl)amino]benzoyl}amino)benzoic acid, ... | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Sreeramulu, S, Nilsson, E, Dekker, N, Wissler, L, Bamberg, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2014-05-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | FXR with DM175 and NCoA-2 peptide
To be Published
|
|
1UJP
 
 | | Crystal Structure of Tryptophan Synthase A-Subunit From Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, Tryptophan synthase alpha chain | | Authors: | Asada, Y, Yokoyama, S, Kuramitsu, S, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-08 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Stabilization mechanism of the tryptophan synthase alpha-subunit from Thermus thermophilus HB8: X-ray crystallographic analysis and calorimetry.
J.Biochem.(Tokyo), 138, 2005
|
|
4QCF
 
 | | Crystal structure of N-terminal mutant (V1A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Alkaline thermostable endoxylanase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2014-05-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into N-terminal to C-terminal interactions and implications for thermostability of a (beta/alpha)8-triosephosphate isomerase barrel enzyme
Febs J., 282, 2015
|
|
4NZY
 
 | | Crystal structure (type-2) of dTMP kinase (st1543) from Sulfolobus Tokodaii Strain7 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into Substrate Binding Mechanism in Thymidylate Kinase based on Intrinsic Dynamics of the Active Site
To be Published
|
|
